1 Introduction
In recent years, deep learning models are being widely used in radiotherapy for the task of medical image segmentation. Although these models have been shown to accelerate clinical workflows (Zabel et al., 2021; Van Dijk et al., 2020), they still commit contouring errors (Brouwer et al., 2020). Thus, a thorough quality assessment (QA) needs to be conducted, which places a higher time and manpower requirement on clinical resources. This creates a barrier to the adoption of such deep learning models (Petragallo et al., 2022). Moreover, it also creates an obstacle for adaptive radiotherapy (ART) workflows, which have been shown to improve a patient’s post-radiation quality-of-life (Grepl et al., 2023). This obstacle arises due to ART’s need of regular contour updates. Currently, commercial auto-contouring tools do not have the ability to assist with quick identification and rectification of potentially erroneous predictions (Brouwer et al., 2020; Petragallo et al., 2022).
Quality assessment (QA) of incorrect contours would require two steps – 1) error detection and 2) error correction (Chaves-de-Plaza et al., 2022). Currently, errors are searched for by manual inspection and then rectified using existing contour editing tools. Error detection could be semi-automated by recommending either potentially erroneous slices of a 3D scan (Wang et al., 2020), or by highlighting portions of the predicted contours (Sander et al., 2020) or blobs (Nair et al., 2020). Upon detection of the erroneous region, the contours could be rectified using point or scribble-based techniques (Lei et al., 2019; Sambaturu et al., 2023) in a manner that adjacent slices are also updated. For this work, we will focus on error detection.
Various approaches to error detection have suggested using Bayesian Deep Learning (BDL) and the uncertainty that it can produce as a method to capture potential errors in the predicted segmentation masks (Wang et al., 2020; Sander et al., 2020; Nair et al., 2020; Garifullin et al., 2021; Bragman et al., 2018; Ng et al., 2022; Camarasa et al., 2021). Although such works established the potential usage of uncertainty in the QA of predictions, it may not be sufficient in a clinical workflow that relies on pixel-wise uncertainty as a proxy for error detection. In our experiments with deep Bayesian models, we observed that the relationship between prediction errors and uncertainty is sub-optimal, and hence has low clinical “utility”. Ideally, for semi-automated contour QA, the uncertainty should be present only in inaccurate regions and not in the accurate ones. At times, literature usually refers to this as uncertainty calibration (Kumar et al., 2019; Krishnan and Tickoo, 2020; Zhang et al., 2020; Camarasa et al., 2021; Gruber and Buettner, 2022), but we find this term incorrect as historically, calibration is referred to in context of probabilities of a particular event (Dawid, 1982). Thus, we believe it is semantically incorrect to say uncertainty calibration and instead propose to use the term uncertainty-error correspondence.
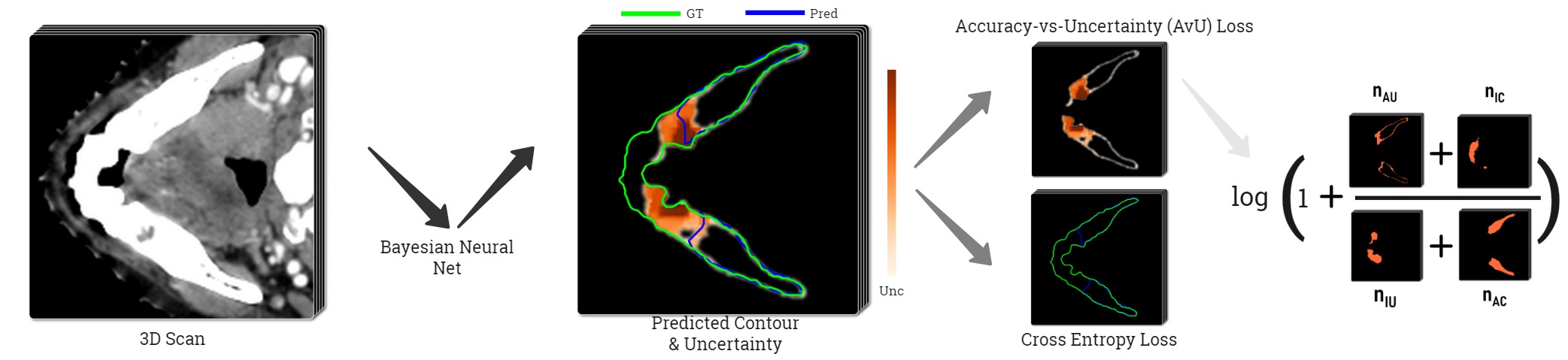
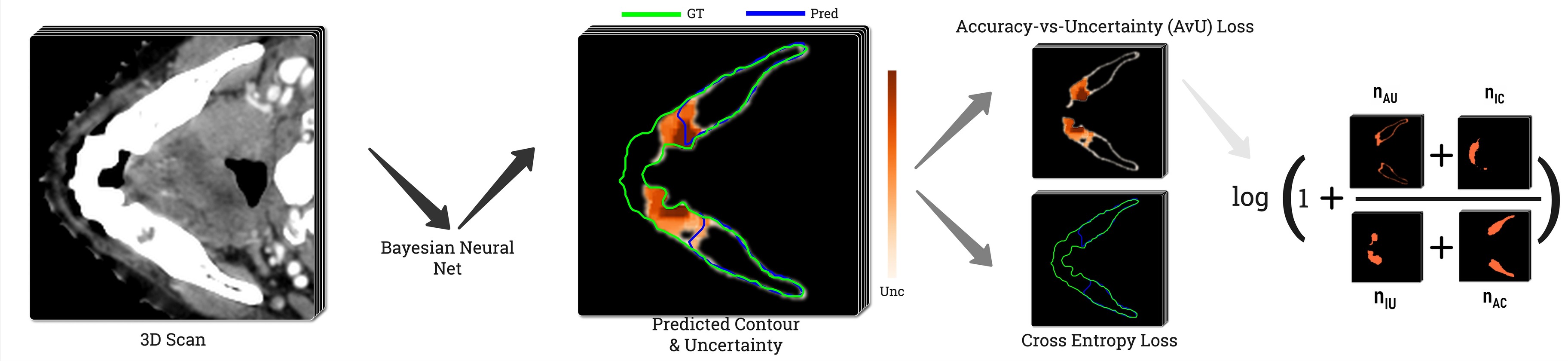
To create a Bayesian model that is incentivized to produce uncertainty only in inaccurate regions, we use the Accuracy-vs-Uncertainty (AvU) metric (Mukhoti and Gal, 2018) and its probabilistic loss version (Krishnan and Tickoo, 2020) during training of a UNet-based Bayesian model (Wen et al., 2018). This loss promotes the presence of both accurate-if-certain () as well as inaccurate-if-uncertain () voxels in the final prediction (Figure 1). With uncertainty present only around potentially inaccurate regions, one can achieve improved synergy between clinical experts and their deep learning tools during the QA stage. Our work is the first to use the AvU loss in a dense prediction task like medical image segmentation and also with datasets containing natural and not synthetic variations as was previously done (Krishnan and Tickoo, 2020). This work extends our conference paper (Mody et al., 2022a) with additional datasets, experiments and metrics. There, we adapt the original AvU loss by considering the full theoretical range of uncertainties in the loss, rather than one extracted from the validation dataset (Krishnan and Tickoo, 2020). For our work we use the predictive entropy as an uncertainty metric (Gal, 2016).
Several other approaches have been considered in context of uncertainty, for e.g. ensembles, test time augmentation (TTA) and model calibration. While ensembles of models have good segmentation performance (Mehrtash et al., 2020; Ng et al., 2022), they are parameter heavy. TTA (Wang et al., 2019b; Hekler et al., 2023) performs inference by modulating a models inputs, but does not perform additional training, so may be unable to transcend its limitations. Calibration techniques attempt to make predictions less overconfident (Guo et al., 2017; Pereyra et al., 2017; Müller et al., 2019; Mukhoti et al., 2020; Islam and Glocker, 2021; Murugesan et al., 2023b), however they do not explicitly align model errors with uncertainty. All the above methods are benchmarked on the truthfulness of their output probabilities (when compared against voxel accuracies) using metrics like expected calibration error (ECE). However, a model with lower ECE than its counterparts may not necessarily have higher uncertainty-error correspondence.
Finally, to evaluate calibrative and uncertainty-error correspondence metrics, one needs to compute the “true” inaccuracy map. Similar to our conference paper (Mody et al., 2022a) and inspired by Sander et al. (2020), we classify inaccuracies of predicted voxel maps into two categories: “errors” and “failures” (see Appendix A). Segmentation “errors” are those inaccuracies which are considered an artifact similar to inter-observer variation, a phenomenon common in medical image segmentation (Brouwer et al., 2012; van der Veen et al., 2019). Thus, we consider these smaller inaccuracies to be accurate in our computations, under the assumption they do not require clinical intervention. In the context of contour QA, such voxels should ideally be certain. Hence, only the segmentation “failures” are a part of the “true” inaccuracy map used to calculate the calibrative and uncertainty-error correspondence metrics.
To summarize, our contributions are as follows:
- •
For the purpose of semi-automated quality assessment of predicted contours, we aim to improve uncertainty-error correspondence (unc-err) in a Bayesian medical image segmentation setting, pioneering this in the context of radiation therapy. Specifically, we propose using the loss form of the Accuracy-vs-Uncertainty (AvU) metric while training a deep Bayesian segmentation model.
- •
We compare our Bayesian model with the AvU loss against an ensemble of deterministic models, five approaches employing calibration-based losses and also test time augmentation. We also perform an architectural comparison by comparing models with Bayesian convolutions placed in either the middle layers or decoder layers of a deep segmentation model.
- •
We benchmark unc-err of the segmentation models on both in- and out-of-distribution radiotherapy datasets containing head-and-neck CT and Prostate MR scans. Models are benchmarked on these datasets across discriminative, calibrative and uncertainty-error correspondence metrics.
2 Related Works
2.1 Epistemic and aleatoric uncertainty
Recent years have seen an increase in work that utilizes probabilistic modeling in deep medical image segmentation. The goal has been to account for uncertainty due to noise in the dataset (aleatoric uncertainty) as well as in the limitations of the predictive models learning capabilities (epistemic uncertainty). Noise in medical image segmentation refers to factors like inter- and intra- annotator contour variation (Brouwer et al., 2012; van der Veen et al., 2019) due to factors such as poor contrast in medical scans. Works investigating aleatoric uncertainty model the contour diversity in a dataset by either placing Gaussian noise assumptions on their output (Monteiro et al., 2020) or by assuming a latent space in the hidden layers and training on datasets containing multiple annotations per scan (Hu et al., 2019). A popular and easy-to-implement approach to model for aleatoric uncertainty is called test-time augmentation (TTA) (Wang et al., 2019a). Here, different transformations of the image are passed through a model, and the resulting outputs are combined to produce both an output and its associated uncertainty.
In contrast to aleatoric uncertainty, epistemic uncertainty could be used to identify scans (or parts of the scan) that are very different from the training dataset. Here, the model is unable to make a proper interpolation from its existing knowledge. Methods such as ensembling (Mehrtash et al., 2020) and Bayesian posterior inference (e.g., Monte-Carlo DropOut, Stochastic Variational Inference) (Sander et al., 2020; Nair et al., 2020; Wang et al., 2020; Garifullin et al., 2021; Bragman et al., 2018; Gibson et al., 2022; Krishnan and Tickoo, 2020) are common methods to model epistemic uncertainty in neural nets. While Bayesian modeling is a more mathematically motivated and hence, principled approach to estimating uncertainty, ensembles have been motivated by the empirically-proven concept of bootstrapping. In contrast to Bayesian models where the perturbation is modelled by placing distributions on weights, ensembles use either different model weight initializations, or different subsets of the training data. In Bayesian inference techniques, perturbations are introduced in the models activation or weight space. Dropout (Gal and Ghahramani, 2016) and DropConnect (Wan et al., 2013) are popular techniques that apply the Bernoulli distribution on these spaces. Stochastic variational inference (SVI) is another type of weight space perturbation that usually assumes the more expressive Gaussian distribution on the weights. Bayes by Backprop (Blundell et al., 2015) and its resource-efficient variant such as FlipOut (Wen et al., 2018) are examples of SVI. For our work, we consider approaches that are designed for both epistemic uncertainty (Ensembles and SVI models) as well as aleatoric uncertainty (TTA).
2.2 Uncertainty use during training
Other works also use the uncertainty from a base segmentation network to automatically refine its output using a follow-up network. This refinement network can be graphical (Soberanis-Mukul et al., 2020) or simply convolutional (Sander et al., 2020). Uncertainty can also be used in an active learning scenario, either with (Diaz-Pinto et al., 2022) or without (Iwamoto et al., 2021) interactive refinement. Shape-based features of uncertainty maps have also been shown to identify false positive predictions (Bhat et al., 2022). Similarly, we too use uncertainty in our training regime, but with the goal of promoting uncertainty only in those regions which are inaccurate, an objective not previously explored in medical image segmentation.
2.3 Model calibration
In context of segmentation, model calibration error is inversely proportional to the alignment of a models output probabilities with its pixel-wise accuracy. Currently there is no proof that reduction in model calibration error leads to improved uncertainty-error correspondence. However, a weak link can be assumed since both are derived from a models output probabilities. It is well known that the probabilities of deterministic models trained on the cross entropy (CE) loss are not well calibrated (Guo et al., 2017). This means that they are overconfident on incorrect predictions and hence fail silently, which is an undesirable trait in context of segmentation QA and needs to be resolved.
To abate this overconfidence issue, methods such as post-training model calibration (or temperature scaling) (Guo et al., 2017; Ding et al., 2021; Ouyang et al., 2022), ensembles (Mehrtash et al., 2020; Ovadia et al., 2019), calibration-focused training losses (Pereyra et al., 2017; Mukhoti et al., 2020; Murugesan et al., 2023b, a) and calibration-focused training targets (Müller et al., 2019; Islam and Glocker, 2021) have been shown to improve model calibration for deterministic models. Temperature scaling, a post-training model calibration technique, has been shown to perform poorly in out-of-domain (OOD) settings (Ovadia et al., 2019), relies wholly on an additional validation dataset and/or needs explicit shape priors (Ouyang et al., 2022).Local temperature scaling techniques have been proposed that calibrate on the image or pixel level (Ding et al., 2021), however they are still conceptually similar to the base method and are hence plagued by the same concerns. Others (Ouyang et al., 2022) used a shape prior module for out-of-domain robustness, but they only introduced synthetic textural variations in their work.
Another approach to model calibration is to regularize a model during train to promote uncertainty. For e.g. the ECP (Pereyra et al., 2017) technique explicitly adds the negative entropy to the training loss. Conversely, the Focal loss (Lin et al., 2017; Mukhoti et al., 2020)attempts to calibrate a model implicitly by assigning lower weights (during training) to more confident predictions. Other methods smooth the hard targets of the ground truth towards a uniform distribution in the limit. For e.g. Label Smoothing (Szegedy et al., 2016; Müller et al., 2019) modifies the class distribution of a pixel by calculating a weighted average (using parameter ) between the hard target and a uniform distribution. On the other hand, Spatial Varying Label Smoothing (SVLS) (Islam and Glocker, 2021) modifies a pixel’s class allocation by considering classes around it. Margin-based Label Smoothing (MBLS) (Liu et al., 2022; Murugesan et al., 2023b) reformulates the above approaches by showing that they essentially perform loss optimization where an equality constraint is applied on a pixels logits. MBLS attempts to achieve the best discriminative-calibrative trade-off by softening this equality constraint. They subtract the max logit of a pixel with its other logits and only penalize those logit distances that are greater than a predetermined margin. Others extend the MBLS framework by either learning class-specific weights for the equality constraint (Liu et al., 2023) or reformulating SVLS to a formulation similar to MBLS (Murugesan et al., 2023a). Although these methods attempt to make models less overconfident, they do not explicitly align a model’s error to its uncertainty.
There also exist other approaches to model calibration for e.g., multi-task learning (Karimi and Gholipour, 2022), mixup augmentation (Thulasidasan et al., 2019) and shape priors (Karimi et al., 2019). Multi-task learning requires additional data that may not always be present, while mixup creates synthetic samples which are not representative of the real data distribution. Finally, shape priors may not be applicable to tumors with variable morphology.
Model calibration techniques are evaluated by metrics like Expected Calibration Error (ECE) and its variants (Nixon et al., 2019), however others have also proposed terms like Uncertainty-Calibration Error (UCE) (Laves et al., 2019; Ghoshal and Tucker, 2022). While ECE evaluates the equivalency between accuracy and predicted probability, UCE compares inaccuracy and uncertainty. However, while it is semantically appropriate to expect an average probability of ) to give the same average accuracy (i.e., the mathematical formulation of ECE), the same is not appropriate for inaccuracy and uncertainty ). Hence, UCE is not applicable to our work.
To conclude, the issue with each of the aforementioned techniques for epistemic, aleatoric and calibrative modeling is that they do not explicitly train the model to develop an innate sense of potential errors on a given segmentation task. Given that this is the primary requirement from a contour QA perspective, these models may be unable to have good uncertainty-error correspondence.
3 Methods
3.1 Neural Architecture
We adopt the OrganNet2.5D neural net architecture (Chen et al., 2021) which is a standard encoder-decoder model connected by four middle layers. It contains both 2D and 3D convolutions in the encoder and decoder as well as hybrid dilated convolutions (HDC) in the middle. This network performs fewer pooling steps to avoid losing image resolution and instead uses HDC to expand the receptive field. To obtain uncertainty corresponding to the output, we add stochasticity to the deterministic convolutional operations by replacing them with Bayesian convolutions (Blundell et al., 2015; Wen et al., 2018). We experiment with replacing deterministic layers in both the HDC as well as the decoder layers to understand the effect of placement.
In a Bayesian model, a prior distribution is placed upon the weights and is then updated to a posterior distribution on the basis of the training data. During inference (Equation 1), we sample from this posterior distribution to estimate the output distribution with , and being the input, output and neural weight respectively:
| (1) |
This work uses a Bayesian posterior estimation technique called stochastic variational inference, where instead of finding the true, albeit intractable posterior, it finds a distribution close to it. We chose FlipOut-based (Wen et al., 2018) convolutions which assume the distribution over the neural weights to be a Gaussian and are factorizable over each hidden layer. Pure variational approaches would need to sample from this distribution for each element of the mini-batch (Blundell et al., 2015). However, the FlipOut technique only samples once and multiplies that random sample with a Rademacher matrix, making the forward pass less computationally expensive.
3.2 Training Objectives
In this section , we use a notation format, where capital letters denote arrays while non-capital letters denote scalar values.
3.2.1 Segmentation Objective
Upon being provided a 3D scan as input, our segmentation model predicts for each class , a 3D probability map of the same size. Each voxel has a predicted probability vector containing values for each class that sum to 1 (due to softmax). To calculate the predicted class of each voxel , we do:
| (2) |
To generate a training signal, the predicted probability vector is compared to the corresponding one-hot vector in the gold standard 3D annotation mask. is composed of . Inspired by (Taghanaki et al., 2019; Yeung et al., 2022), we re-frame the binary cross-entropy loss (Equation 3), as penalizing both the foreground () and background (() voxels of the probability maps of each class with a weight :
| (3) |
Note, we do not utilize the DICE loss for training as it has been shown to have lower model calibration metrics (Mody et al., 2022b). Also, since the CE loss is susceptible to fail during a class-imbalance, we use its weighted version.
3.2.2 Uncertainty Objective
In a Bayesian model, multiple forward passes () are performed and the output 3D probability maps of each pass are averaged to output (Equation 1). Using , we can calculate a host of statistical measures like entropy, mutual information and variance. We chose entropy as it has been shown to capture both epistemic uncertainty, which we explicitly model in FlipOut layers, as well as aleatoric uncertainty, which is implicitly modeled due to training data (Gal, 2016). We use the predicted class probability vector for each voxel and calculate its (normalized) entropy :
| (4) |
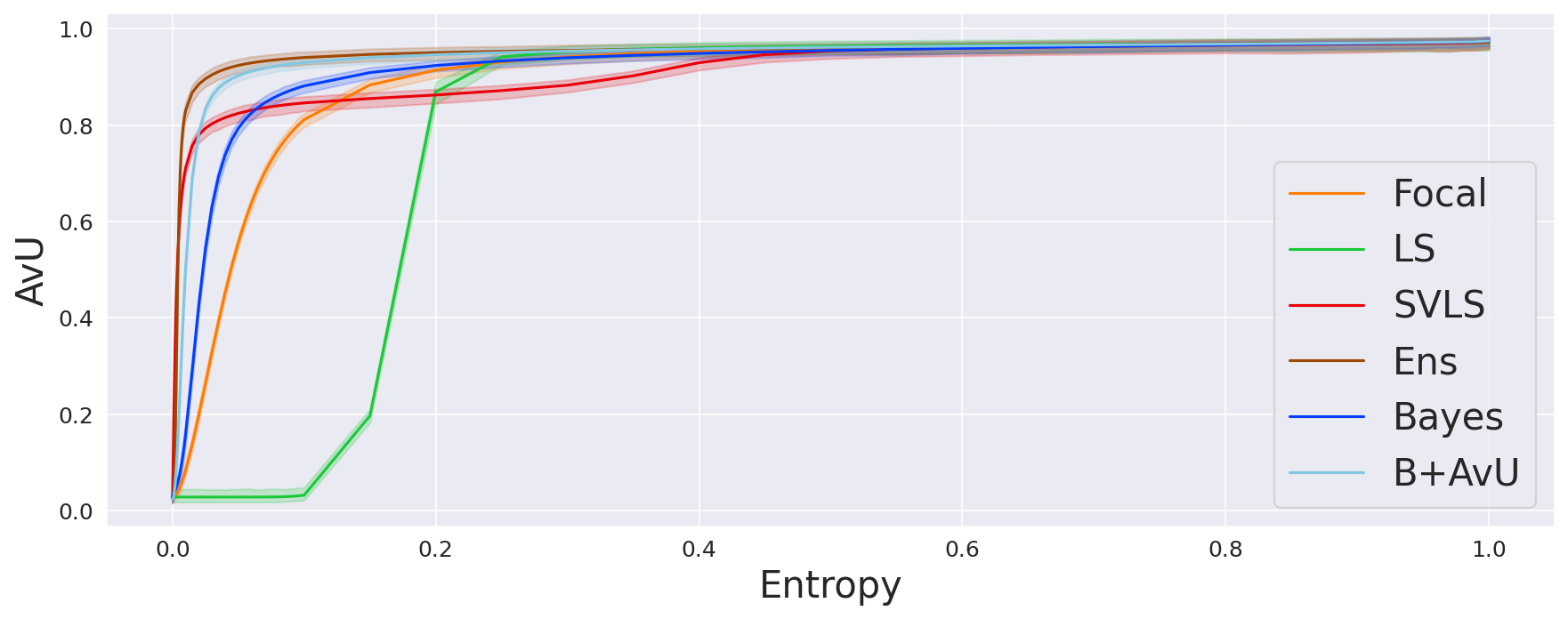
Since we have access to the gold standard annotation mask, each voxel has two properties: accuracy and uncertainty. Accuracy is determined by comparing the gold standard class to the predicted class . We use this to classify them in four different categories represented by , , and , where stands for the total voxel count and , , , represent the accurate, inaccurate, uncertain and certain voxels. A visual representation of these terms can be seen in Figure 1. Here, a voxel is determined to be certain or uncertain on the basis of a chosen uncertainty threshold where the maximum value in is the maximum theoretical uncertainty threshold (Mody et al., 2022a). The aforementioned four terms are the building blocks of the Accuracy-vs-Uncertainty (AvU) metric (Mukhoti and Gal, 2018) as shown in Equation 5 - Equation 7 and it has a range between [0,1]. A higher value indicates that uncertainty is present less in accurate regions and more in inaccurate regions, thus improving the “utility” of uncertainty as a proxy for error detection.
| (5) | ||||
| (6) | ||||
| (7) |
To maximize AvU for a neural net, one can turn it into a loss metric to be minimized. As done in Krishnan and Tickoo (2020) for an image classification setting, we minimize its negative logarithm (Equation 8) to improve mathematical stability of gradient descent. However, the AvU metric, as defined above, is not differentiable with respect to the neural net’s weights. This is due to all its constituent terms being produced either due to thresholding or max operations which introduce discontinuities that disrupt gradient flows.. The AvU metric is made differentiable by instead using the uncertainty derived from (Equation 4), thus allowing for gradient flows. . Also, a smooth non-linear operation i.e., tanh is used to constrain its value (Equation 9). The differentiable uncertainty term is multiplied by other scalar weighing terms c.f. the maximum probability () and accuracy/inaccuracy mask for a voxel. All these operations together allow us to calculate proxy values for , , and . In addition, rather than evaluating the loss at a single uncertainty threshold, we integrate over the theoretical range of the uncertainty metric. Thresholding is done by once again multiplying the uncertainty value with a binary mask. The benefits of thresholding were shown in our conference paper (Mody et al., 2022a):
| (8) | ||||
where
| (9) |
Finally, the total loss combines the segmentation and uncertainty loss as:
| (10) |
3.3 Evaluation
3.3.1 Discriminative and Calibration Evaluation
We evaluate all models on the DICE coefficient for discriminative performance. Calibration is evaluated using the Expected Calibration Error (ECE) (Guo et al., 2017). Numerical results are compared with the Wilcoxon signed-ranked test where a p-value 0.05 is considered significant.
3.3.2 Uncertainty Evaluation
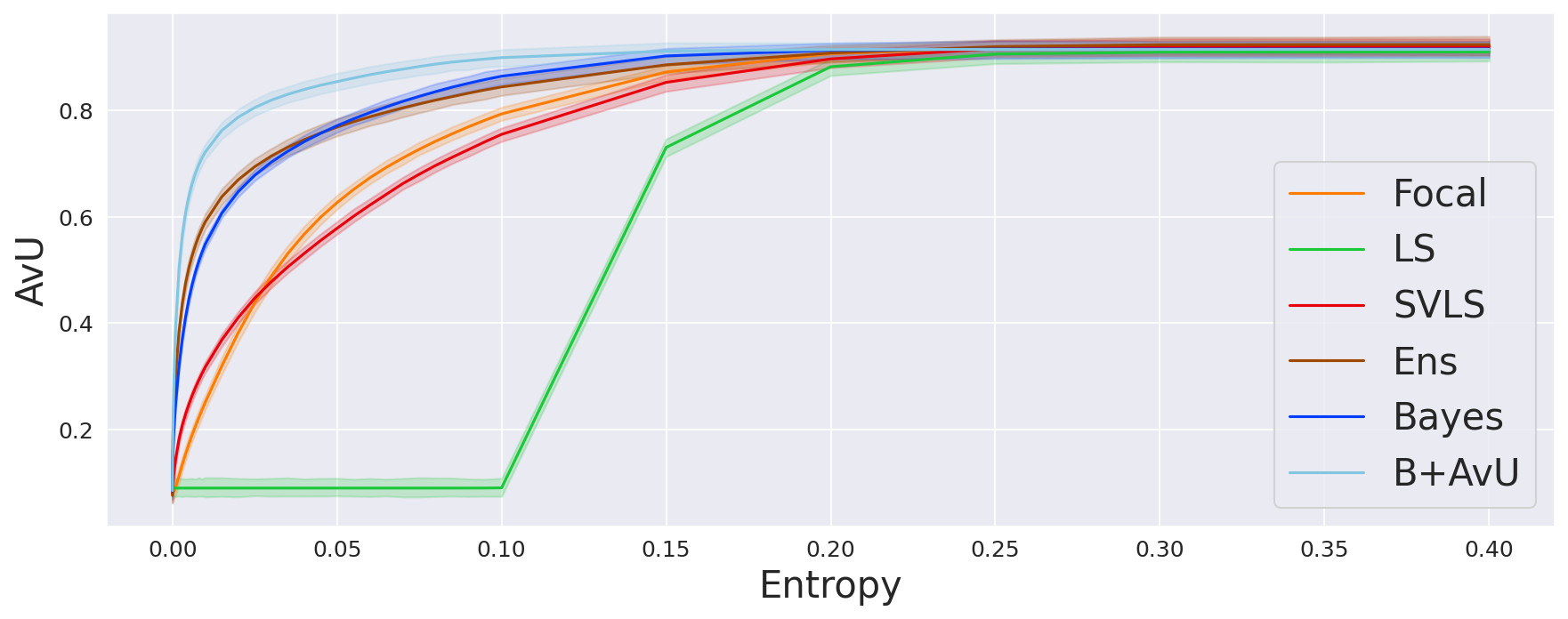
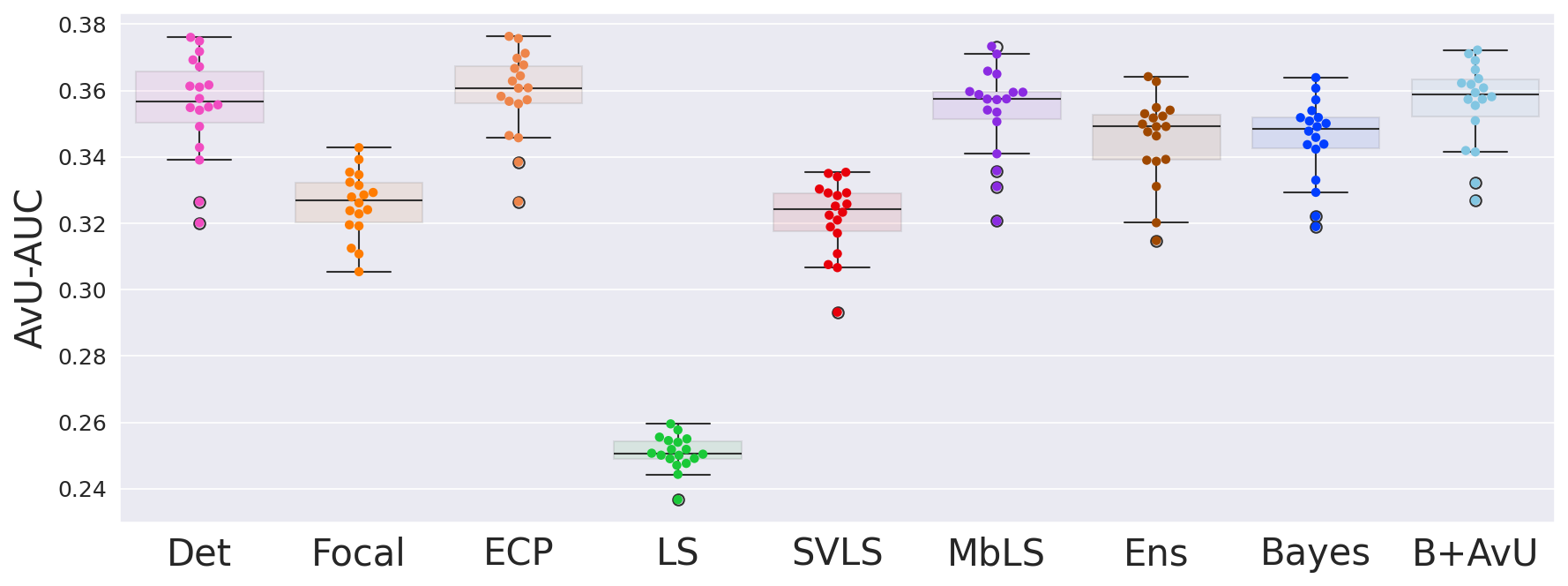
As the model is trained on the Accuracy-vs-Uncertainty (AvU) metric, we calculate the AvU scores up to the maximum normalized uncertainty of the validation dataset. A curve with the AvU score on the y-axis and the uncertainty threshold on the x-axis is made and the area-under-the-curve (AUC) for each scan is calculated. AUC scores provide us with a summary of the model performance regardless of the uncertainty threshold, and hence we use it to compare all models.
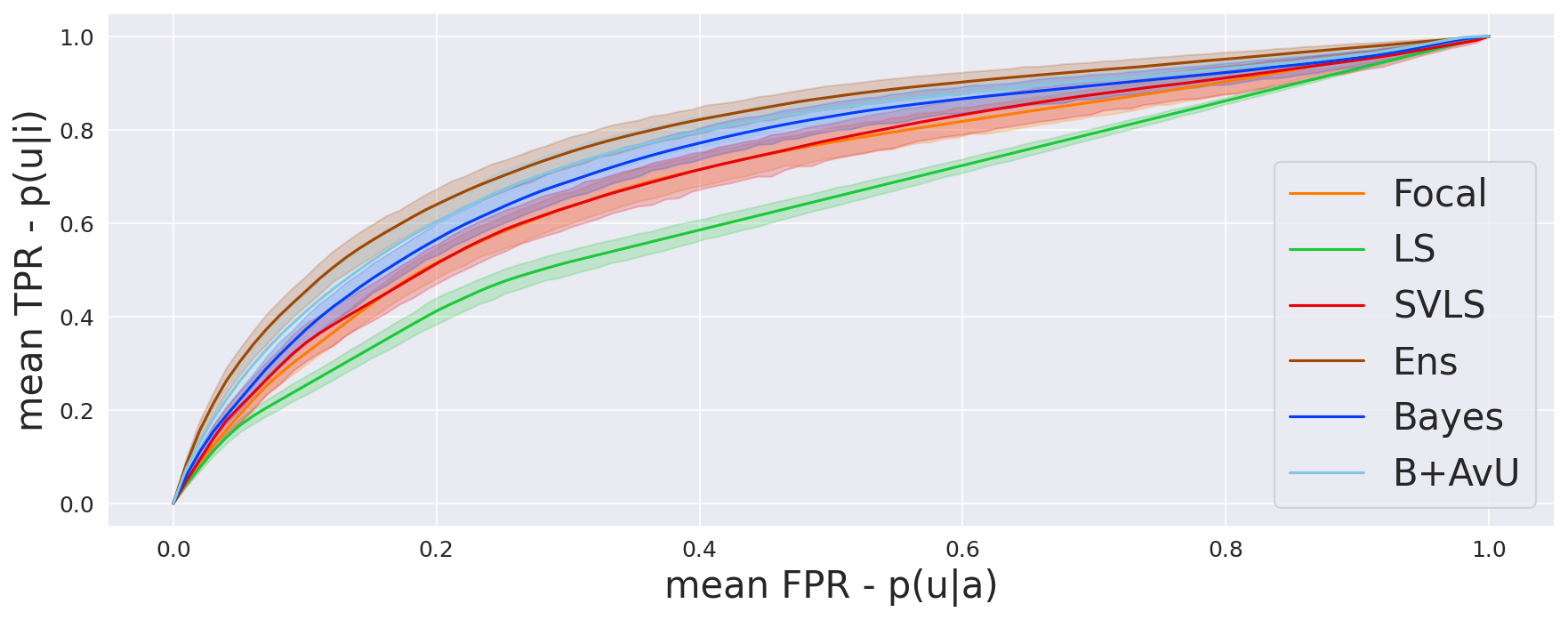
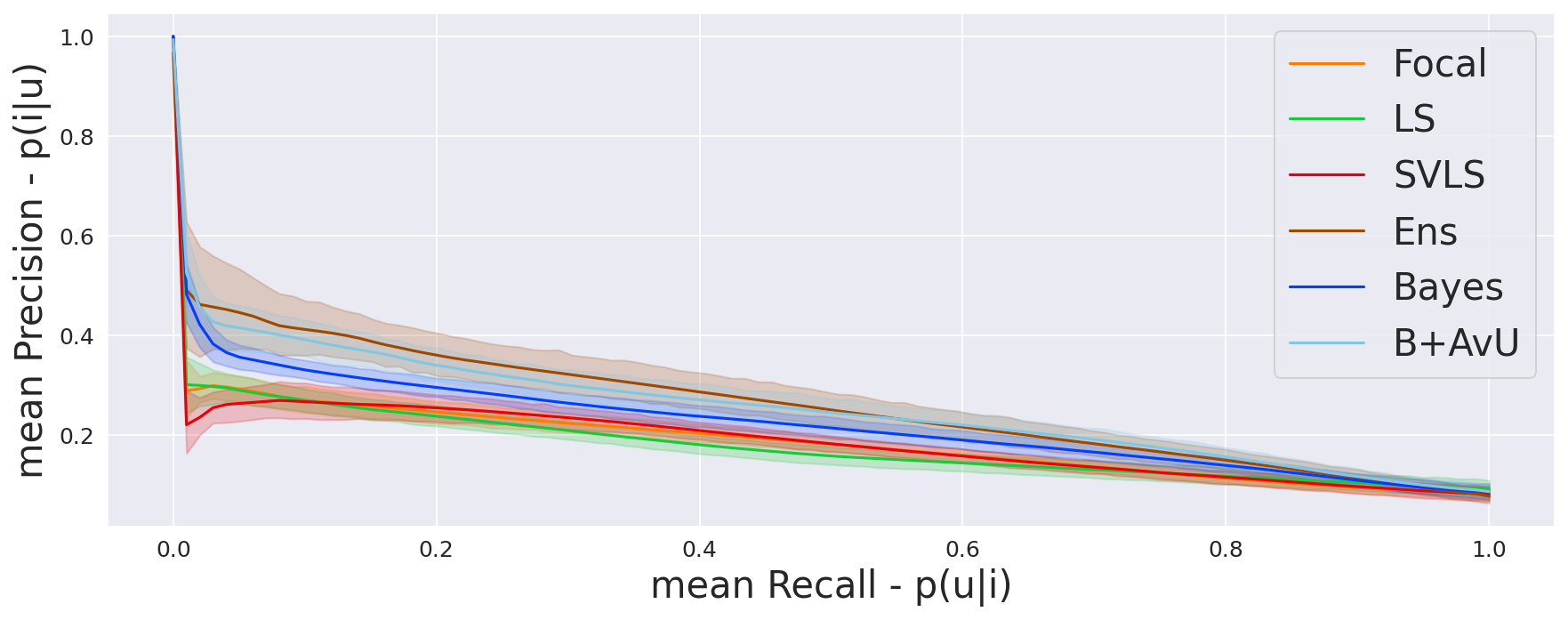
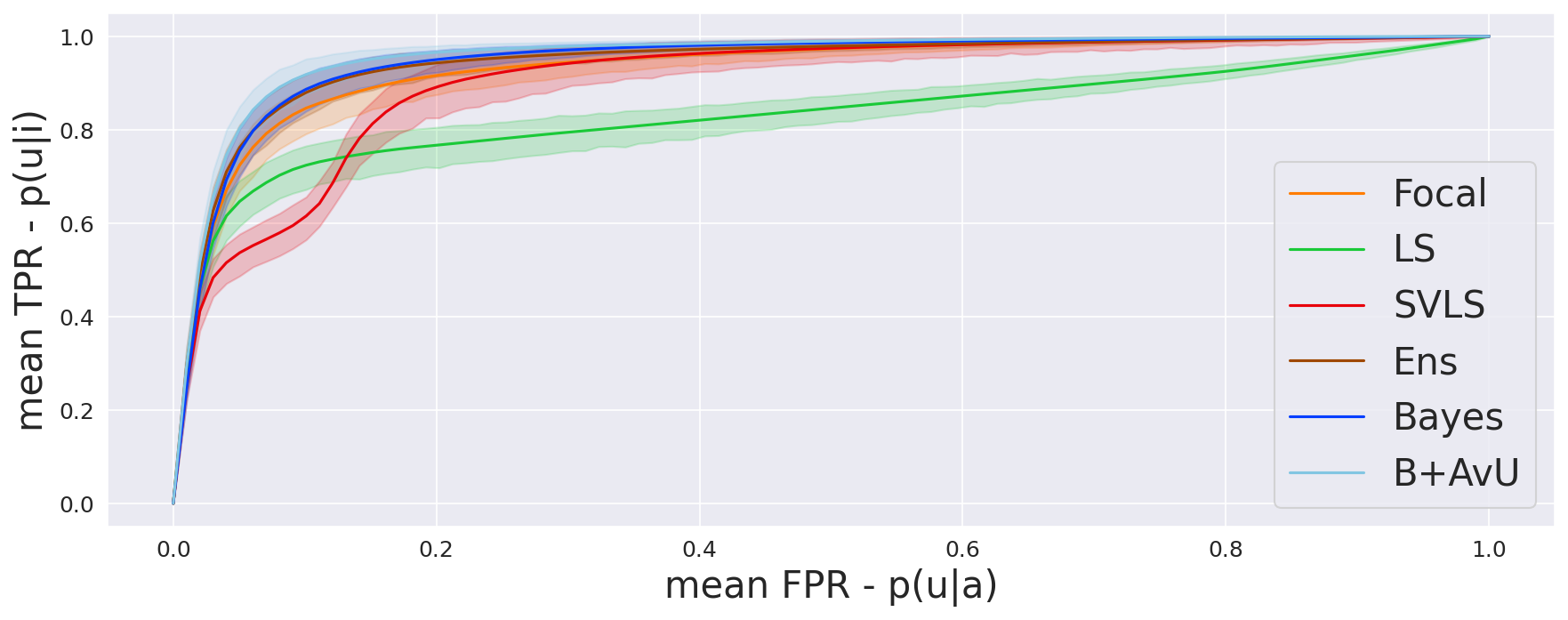
The AvU metric outputs a single scalar value for the whole scan and does not offer much insight into the differences in uncertainty coverage between the accurate and inaccurate regions. To abate this, we compare the probability of uncertainty in inaccurate regions to the probability of uncertainty in accurate regions . Let us plot and on the y-axis and x-axis of a graph respectively, and define , , and , as the count of true positives, false positives, true negatives and false negatives respectively. Thus, is the true positive rate and is the false positive rate. Computing this at different uncertainty thresholds provides us with the Receiver Operating Characteristic (ROC) curve, which we call the uncertainty-ROC curve (Wan et al., 2013).
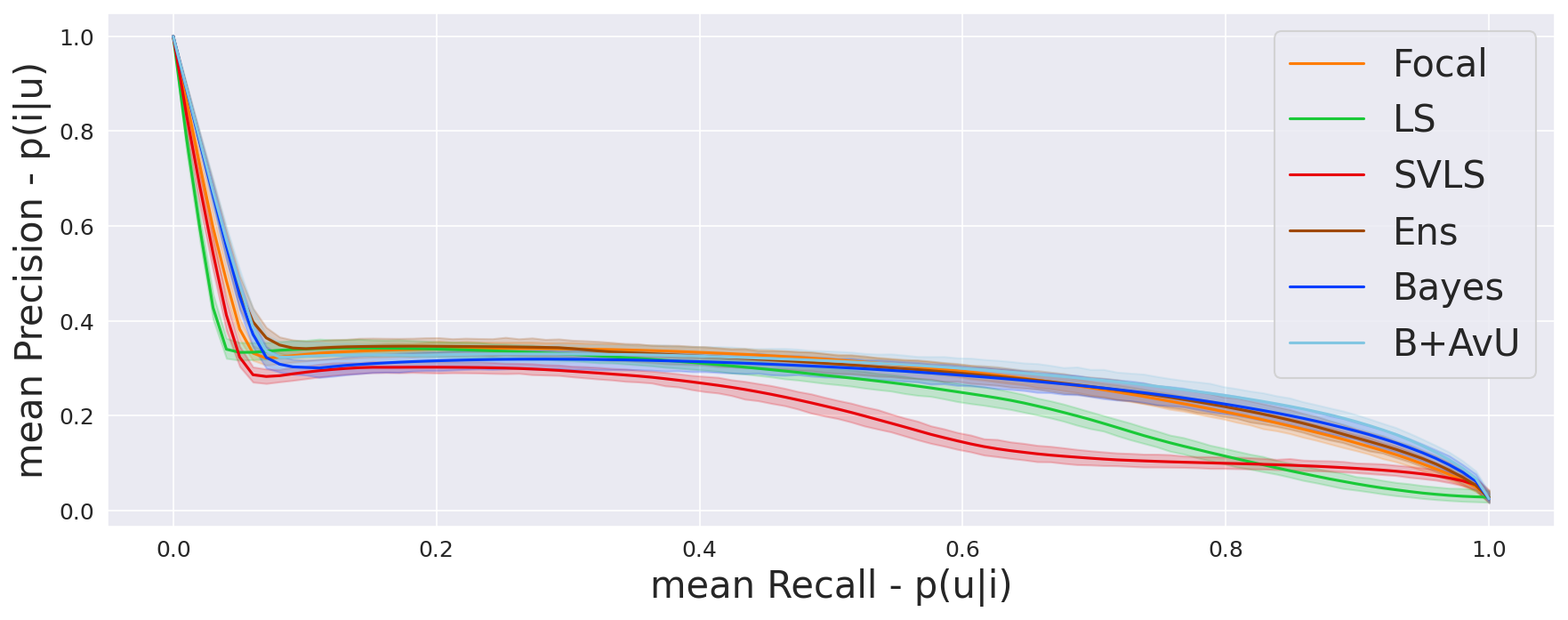
Given that ROC curves are biased in situations with class imbalances between positive (inaccurate voxels) and negative (accurate voxels) classes, we also compute the precision-recall curves (Camarasa et al., 2021).Here, precision is the probability of inaccuracy given uncertainty and recall is the probability of uncertainty given inaccuracy . Note, that the precision-recall curves do not make use of , which can be high in count for a well-performing model.
Finally, to calculate the calibrative and uncertainty-correspondence metrics, we need an inaccuracy map. We use an inaccuracy map based on the concept of segmentation “failures” and “errors” (Appendix A). To do this, we perform a morphological opening operation using a fixed kernel size of (3,3,1).
4 Experiments and Results
4.1 Datasets
4.1.1 Head-and-Neck CT
Our first dataset contained Head and Neck CT scans of patients from the RTOG 0522 clinical trial (Ang et al., 2014). The annotated data, which had been collected from the MICCAI2015 Head and Neck Segmentation challenge, contained 33 CT scans for training, 5 for validation and 10 for testing (Raudaschl et al., 2017). We further expanded the test dataset with annotations of 8 patients belonging to the RTOG trial from the DeepMindTCIA dataset (DTCIA) (Nikolov et al., 2021). This dataset included annotations for the mandible, parotid glands, submandibular glands and brainstem. Although there were annotations present for the optic organs, we ignored them for this analysis as they are smaller compared to other organs and require special architectural design choices. Since the train and test patients came from the same study, we considered this as an in-distribution dataset. We also tested our models on the STRUCTSeg (50 scans) dataset (Ye et al., 2022), hereby shortened as STRSeg. While the RTOG dataset contained American patients, the STRSeg dataset was made up of Chinese patients and hence considered out-of-distribution (OOD) in context of the training data. The uncertainties of this dataset were evaluated to a value of 0.4 since that is the maximum empirical normalized entropy.
4.1.2 Prostate MR
Our second dataset contained MR scans of the prostate for which we use the ProstateX repository (Meyer et al., 2021) containing 66 scans as the training dataset. The Medical Decathlon (Prostate) dataset with 34 scans (Antonelli et al., 2022) and the PROMISE12 repository with 50 scans (Litjens et al., 2014) served as our test dataset. The Medical Decathlon dataset (abbreviated as PrMedDec henceforth) contained scans from the same clinic as the ProstateX training dataset. We combined the Peripheral Zone (PZ) and Transition Zone (TZ) from the MedDec dataset into 1 segmentation mask. The PROMISE12 dataset (abbreviated as PR12) was chosen for testing since literature (Mehrtash et al., 2020) has shown lower performance on it and hence it serves as a good candidate to evaluate the utility of uncertainty. This dataset is different from ProstateX due to the usage of an endo-rectal coil in many of its scans as well as the presence of gas pockets in the rectum and dark shadows due to the usage of older MR machines. Thus, although these datasets contained scans of the prostate region, there exists a substantial difference in their visual textures. The maximum empirical normalized entropy of this 2-class dataset is 1.0 and hence the uncertainty-error correspondence metrics were calculated till this value.
4.2 Experimental Settings
We tested the Accuracy-vs-Uncertainty (AvU) loss on four datasets containing scans of different modalities and body sites. We trained 11 models: Det (deterministic), Det+AvU, Ensemble, Focal, LS (Label Smoothing), SVLS (Spatially Varying Label Smoothing), MbLS (Margin based Label Smoothing), ECP (Explicit Confidence Penalty), TTA (Test-Time Augmentation), Bayes and Bayes + AvU. As the names suggest, Bayes and Bayes + AvU are Bayesian versions of the deterministic OrganNet2.5D model (Chen et al., 2021). The baseline Bayes model contained Bayesian convolutions in its middle layers and was trained using only the cross-entropy (CE) loss. The Bayes + AvU was trained using both the CE and Accuracy-vs-Uncertainty (AvU) loss. Two additional Bayesian models were trained which tests if the placement of the Bayesian layers had any effect: BayesH and BayesH + AvU. Here, BayesH refers to the Bayesian model with Bayesian layers in the head of the model (i.e the decoder). Results for these models can be found in Appendix E.
The Ensemble was made of deterministic models with different initializations (Ovadia et al., 2019). For TTA, we applied Gaussian noise and random pixel removals for times each and then averaged their outputs. The hyperparameters of the other models were chosen on the basis of the best discriminative, calibrative and uncertainty-error correspondence metrics on the validation datasets (C). For the calibration focused methods we used the following range of hyperparameters: Focal (), MbLS () for head-and-neck CT, MbLS () for prostate MR, LS (), SVLS () and ECP () for head-and-neck CT and ECP () for prostate MR. For the AvU loss, we evaluated weighting factors in the range 10,100,1000,10000 for the head-and-neck dataset, and 100,1000,10000 for the Prostate dataset.
We trained our models for 1000 epochs using the Adam optimizer with a fixed learning rate of 10-3. The deterministic model contained 550K parameters and thus the Ensemble contained 2.75M parameters. Since the Bayesian models double the parameter count in their layers they incurred an additional parameter cost and ended up with a total of 900K parameters.
4.3 Results
In Section 4.3.1 and Section 4.3.2 we show discriminative (DICE), calibrative (ECE) and uncertainty-error correspondence metrics (ROC-AUC, PRC-AUC) for the two datasets.
4.3.1 Head-and-neck CT
| TestDataset | Model | DICE (x) | ECE (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| ID————H&N CT(RTOG) | Det | 84.2 ± 2.7 | 9.0 ± 2.1 | 73.0 ± 5.7 | 21.0 ± 4.8 |
| Det + AvU | 83.8 ± 2.9 | 8.6 ± 2.7 | 73.1 ± 6.0 | 20.8 ± 4.0 | |
| 2-6 | Focal | 84.3 ± 2.4 | 9.3 ± 1.5 | 70.3 ± 5.5 | 18.2 ± 3.2 |
| ECP | 84.4 ± 2.3 | 9.0 ± 2.0 | 73.8 ± 5.4 | 21.0 ± 3.7 | |
| LS | 83.0 ± 3.0 | 7.5 ± 2.2 | 62.6 ± 3.3 | 17.5 ± 4.0 | |
| SVLS | 84.2 ± 2.6 | 9.0 ± 2.0 | 70.8 ± 7.1 | 18.1 ± 3.5 | |
| MbLS | 84.0 ± 2.6 | 9.2 ± 2.1 | 67.5 ± 5.7 | 19.5 ± 3.5 | |
| 2-6 | TTA | 84.1 ± 2.8 | 9.1 ± 2.1 | 72.9 ± 5.9 | 20.8 ± 3.9 |
| 2-6 | Ensemble | 85.0 ± 2.6 | 7.8 ± 1.8 | 78.6 ± 4.7 | 25.7 ± 6.8 |
| 2-6 | Bayes | 83.9 ± 2.6 | 8.6 ± 2.1 | 74.1 ± 5.4 | 22.1 ± 3.5 |
| Bayes+AvU | 83.6 ± 2.5 | 7.6 ± 2.5† | 76.1 ± 5.6† | 25.1 ± 5.3† | |
| OOD————H&N CT(STRSeg) | Det | 78.1 ± 4.6 | 12.9 ± 2.6 | 62.2 ± 4.5 | 24.1 ± 3.7 |
| Det + AvU | 78.6 ± 4.7 | 12.7 ± 3.0 | 60.8 ± 4.7 | 22.4 ± 4.1 | |
| 2-6 | Focal | 77.2 ± 6.7 | 12.5 ± 2.9 | 57.0 ± 4.6 | 20.9 ± 4.2 |
| ECP | 78.8 ± 4.3 | 12.5 ± 2.6 | 61.5 ± 4.8 | 23.2 ± 3.6 | |
| LS | 77.7 ± 6.0 | 10.3 ± 2.9 | 56.7 ± 3.3 | 20.6 ± 4.3 | |
| SVLS | 79.0 ± 6.0 | 11.3 ± 2.5 | 59.9 ± 5.4 | 21.6 ± 2.7 | |
| MbLS | 77.5 ± 6.3 | 13.4 ± 3.0 | 56.9 ± 5.0 | 21.5 ± 3.6 | |
| 2-6 | TTA | 78.1 ± 4.6 | 12.7 ± 2.6 | 62.7 ± 4.6 | 24.9 ± 4.1 |
| 2-6 | Ensemble | 78.6 ± 5.2 | 10.6 ± 2.4 | 64.7 ± 4.9 | 28.2 ± 5.1 |
| 2-6 | Bayes | 75.0 ± 9.9 | 12.4 ± 4.0 | 64.8 ± 5.0 | 27.7 ± 5.8 |
| Bayes+AvU | 76.3 ± 7.7 | 12.1 ± 3.7 | 65.8 ± 5.0† | 30.1 ± 6.5† |
| TestDataset | Model | DICE (x) | ECE (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| ID————Prostate MR(PrMedDec) | Det | 84.1 ± 5.6 | 12.9 ± 6.0 | 92.5 ± 5.7 | 28.0 ± 3.7 |
| Det + AvU | 83.7 ± 6.8 | 16.9 ± 8.1 | 92.1 ± 6.8 | 28.2 ± 3.4 | |
| 2-6 | Focal | 81.1 ± 15.4 | 10.2 ± 5.0 | 93.2 ± 5.5 | 29.3 ± 3.4 |
| ECP | 84.0 ± 5.5 | 16.7 ± 7.1 | 92.1 ± 6.0 | 27.6 ± 4.3 | |
| LS | 83.4 ± 7.2 | 15.1 ± 8.6 | 83.2 ± 7.8 | 25.1 ± 3.1 | |
| SVLS | 83.5 ± 6.7 | 14.0 ± 8.1 | 90.5 ± 7.9 | 21.7 ± 2.6 | |
| MbLS | 84.2 ± 4.9 | 17.9 ± 7.4 | 92.2 ± 5.6 | 26.9 ± 3.6 | |
| 2-6 | TTA | 83.8 ± 5.8 | 16.4 ± 7.1 | 92.7 ± 5.6 | 28.8 ± 3.9 |
| 2-6 | Ensemble | 84.5 ± 5.7 | 11.3 ± 6.5 | 94.3 ± 4.3 | 30.0 ± 4.6 |
| 2-6 | Bayes | 84.0 ± 5.8 | 8.6 ± 4.7 | 94.7 ± 3.1 | 29.1 ± 4.8 |
| Bayes+AvU | 84.9 ± 6.9 | 8.9 ± 6.0 | 95.7 ± 3.2† | 30.5 ± 4.5† | |
| OOD————Prostate MR(PR12) | Det | 74.2 ± 12.6 | 15.6 ± 6.3 | 87.9 ± 7.5 | 22.1 ± 6.2 |
| Det + AvU | 74.5 ± 13.0 | 27.6 ± 14.3 | 88.2 ± 7.6 | 22.0 ± 7.1 | |
| 2-6 | Focal | 71.2 ± 17.4 | 12.1 ± 5.8 | 89.0 ± 7.1 | 24.3 ± 6.7 |
| ECP | 74.8 ± 12.5 | 22.3 ± 10.2 | 87.2 ± 8.1 | 20.6 ± 7.0 | |
| LS | 74.5 ± 13.0 | 21.7 ± 11.5 | 79.5 ± 8.9 | 19.1 ± 7.2 | |
| SVLS | 76.9 ± 11.5 | 17.9 ± 9.3 | 87.2 ± 7.2 | 16.4 ± 5.2 | |
| MbLS | 73.6 ± 12.5 | 19.9 ± 7.4 | 86.5 ± 7.2 | 21.8 ± 5.6 | |
| 2-6 | TTA | 74.0 ± 12.8 | 23.7 ± 11.4 | 88.6 ± 7.4 | 24.9 ± 5.8 |
| 2-6 | Ensemble | 76.3 ± 12.2 | 9.7 ± 5.0 | 91.6 ± 5.2 | 28.4 ± 5.7 |
| 2-6 | Bayes | 70.6 ± 16.6 | 11.8 ± 7.2 | 89.1 ± 7.4 | 25.7 ± 5.1 |
| Bayes+AvU | 76.3 ± 12.6 | 11.4 ± 6.7 | 90.6 ± 6.9† | 26.2 ± 7.4† |
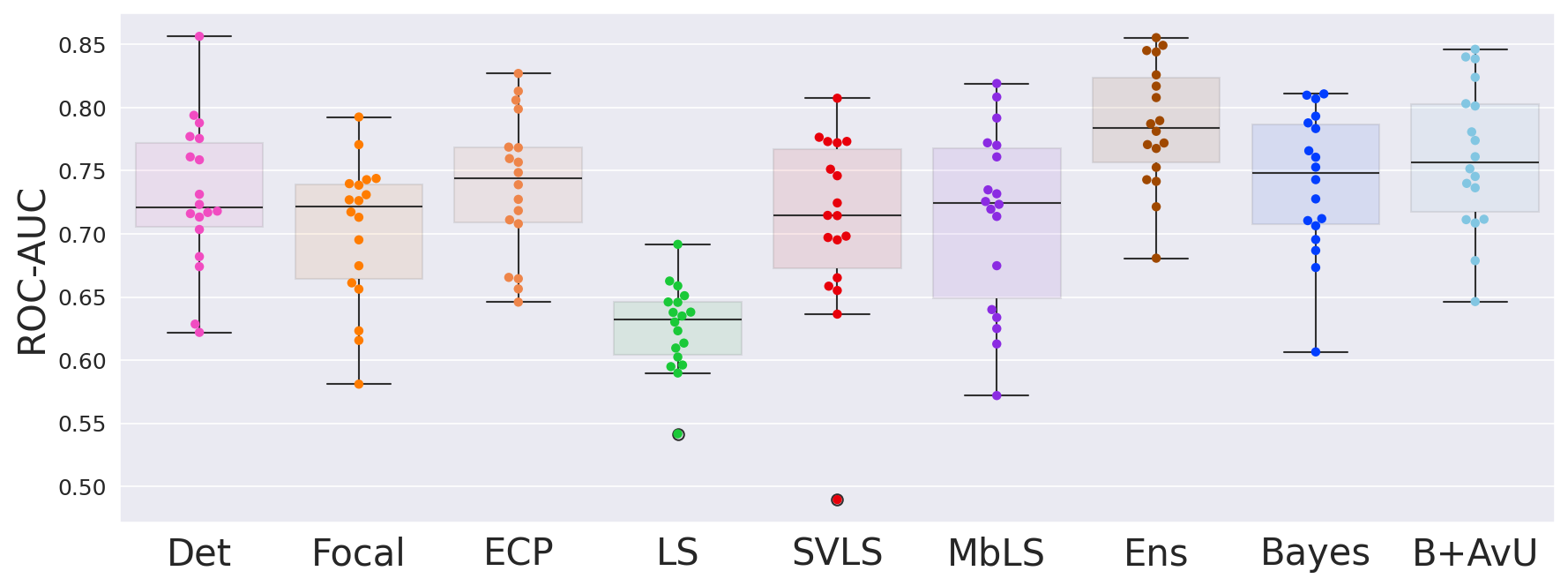
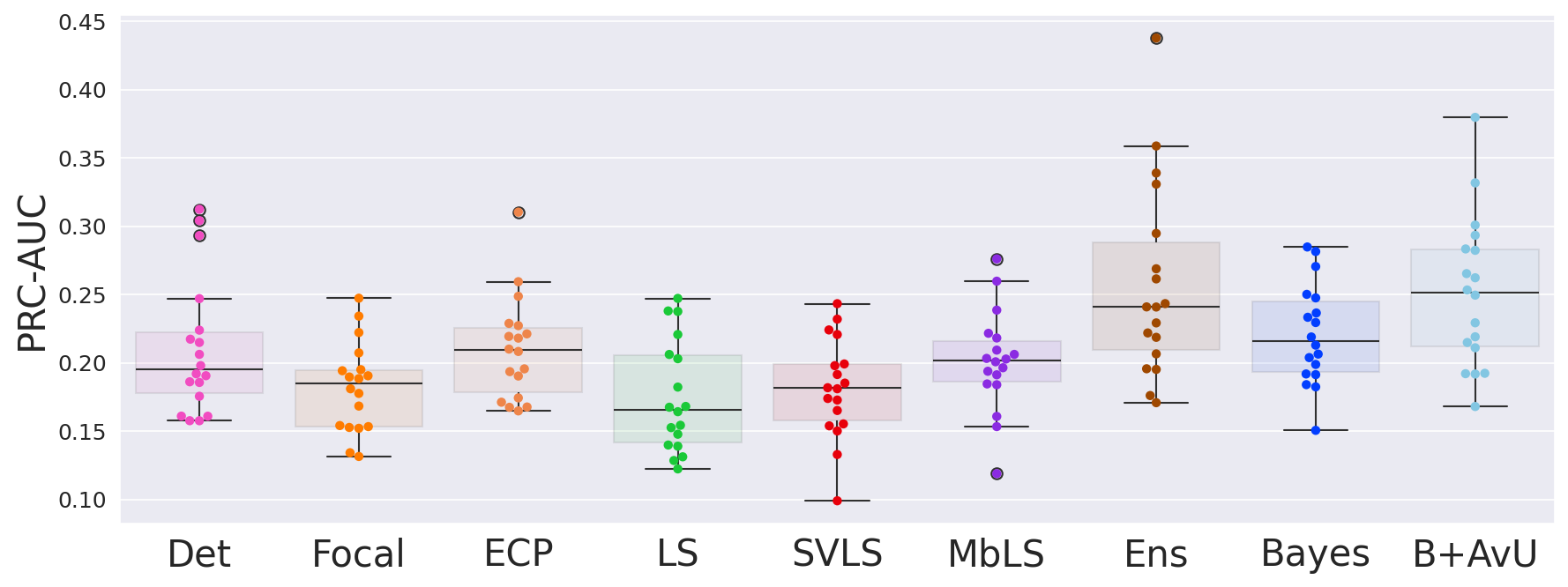
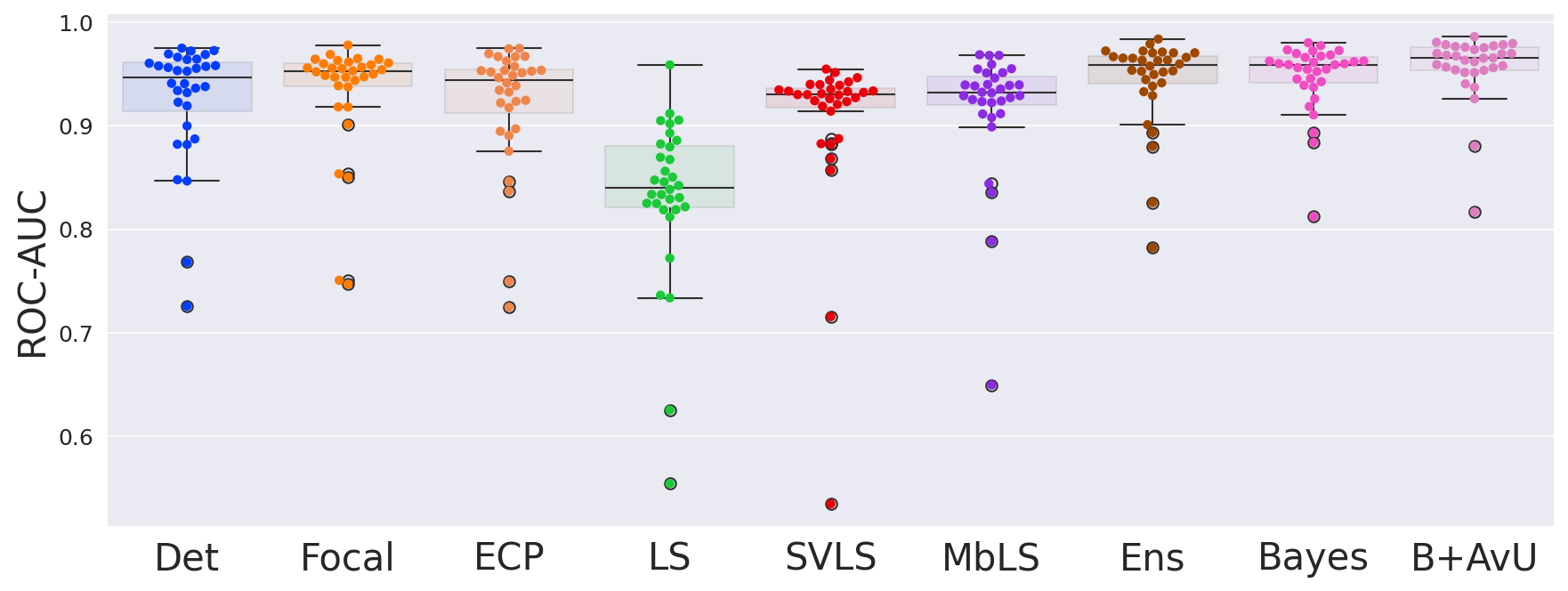
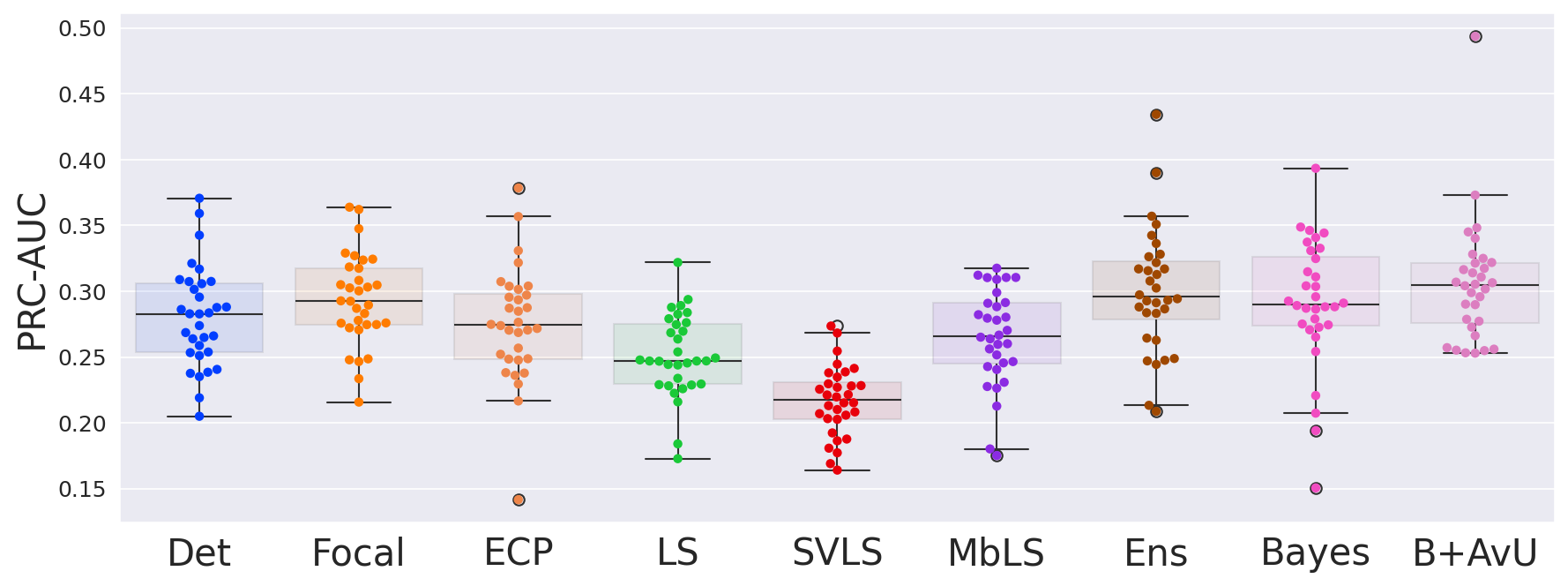
Results in Table 1 showed that the AvU loss on the Bayes model significantly improved calibrative and uncertainty-error correspondence (unc-err) metrics for both in-distribution (ID) and out-of-distribution (OOD) datasets. The Bayes+AvU model also always performed better than the Det, calibration-focused and TTA models for unc-err metrics. Also, its ECE scores were in most cases better than calibration-focused models. However, there was no clear distinction between the performance of the Ensemble and Bayes+AvU model for ECE and unc-err metrics across both datasets. Also, the AvU loss did not benefit the unc-err metrics for the Det model, in both datasets. Of all the calibration-focused models, LS had the lowest ECE and unc-err metrics, while the ECP model had the best unc-err metrics. When compared to Det, the TTA model improved calibrative and unc-err metrics for the OOD dataset, while maintaining it for the ID dataset.
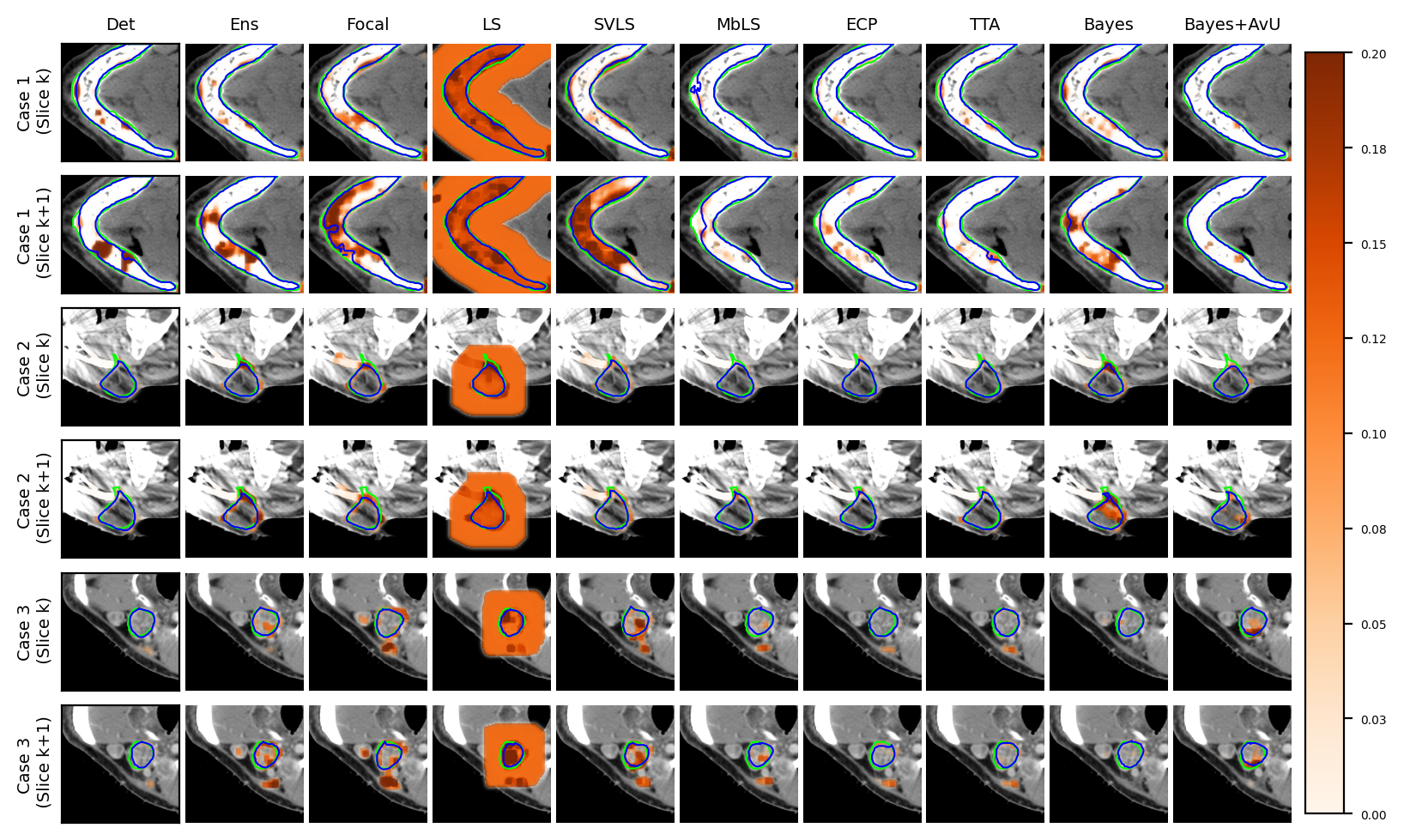
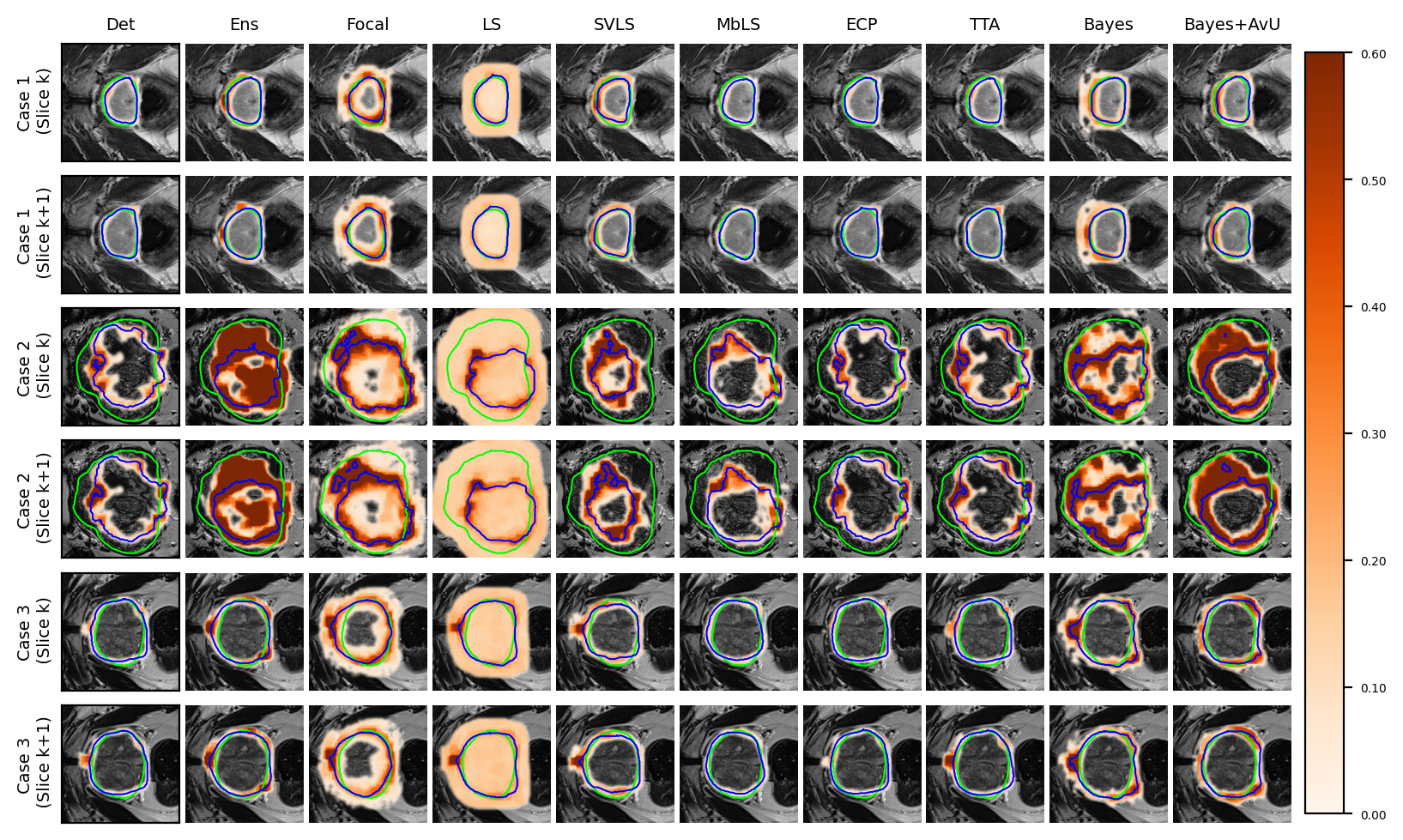
Visually, the Bayes+AvU model was able to successfully suppress uncertainty in the true positive (TP) (Case 1/2 in Figure 2a) and true negative (TN) (Case 3 in Figure 2a) regions of the predicted contour. Moreover, it also showed uncertainty in false positive (FP) regions while also suppressing uncertainty in TP regions (Case 3 in Figure 2b). Calibrative models (e.g. Focal, LS, SVLS) tended to be quite uncertain in TP or TN regions, which may lead to additional QA time. Detailed descriptions are provided in Appendix D.
4.3.2 Prostate MR
Similar to the head-and-neck CT dataset, the use of the AvU loss on the baseline Bayes model significantly improved its uncertainty-error correspondence (unc-err) while maintaining calibration performance (Table 2). Moreover, it improved the DICE values such that its one of the most competitive amongst all models. Also, the Bayes+AvU had better performance in both unc-err and calibrative metrics when compared to the Det, calibration-focused and TTA models. When comparing to the Ensemble, the Bayes+AvU had similar DICE. While Bayes+AvU had better calibrative and unc-err performance in the in-distribution (ID) dataset, the Ensemble performed better in the out-of-distribution (OOD) setting. The AvU loss had no positive effect on the DICE and unc-err performance of the Det model in both the ID and OOD setting, however there was an increase in ECE.
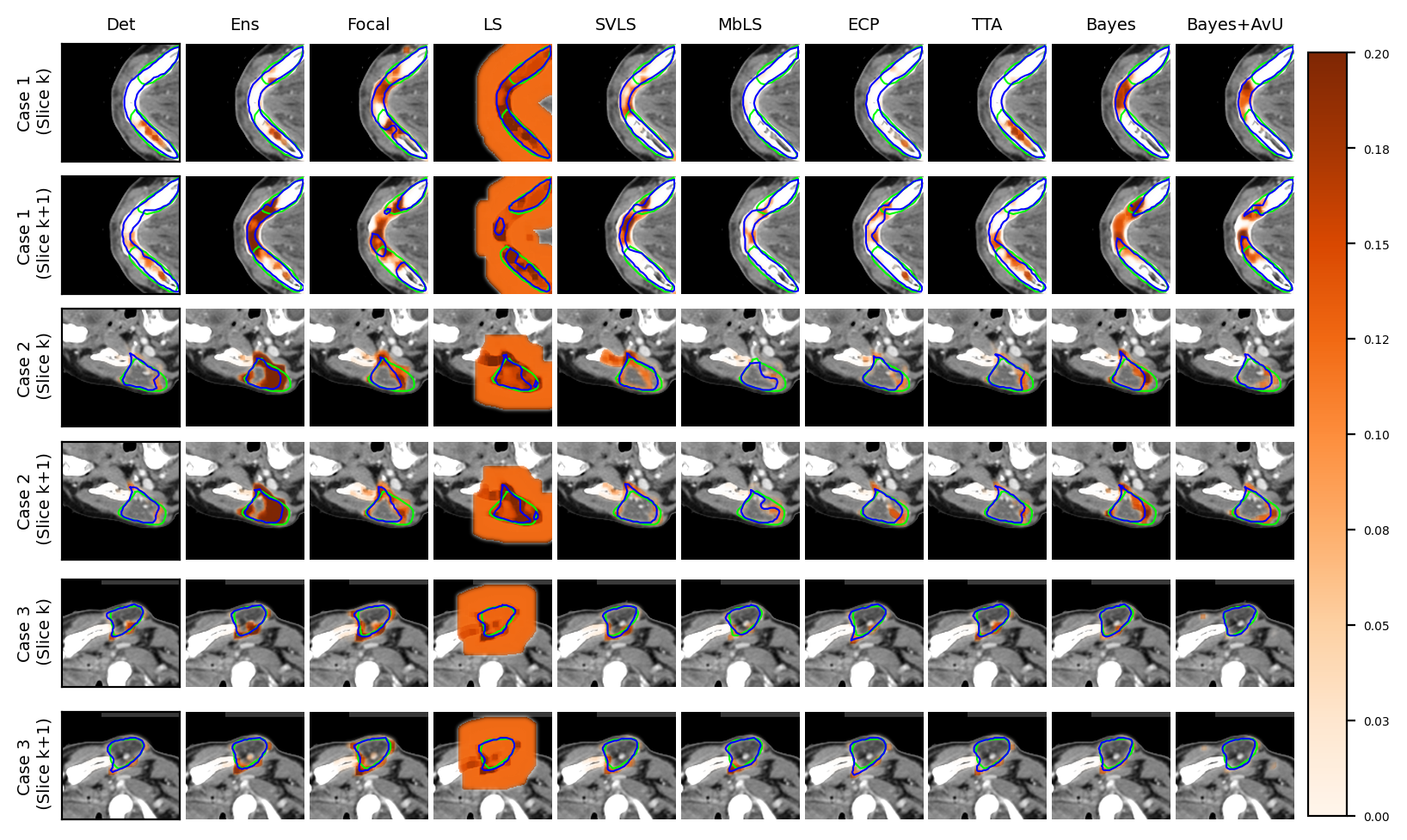
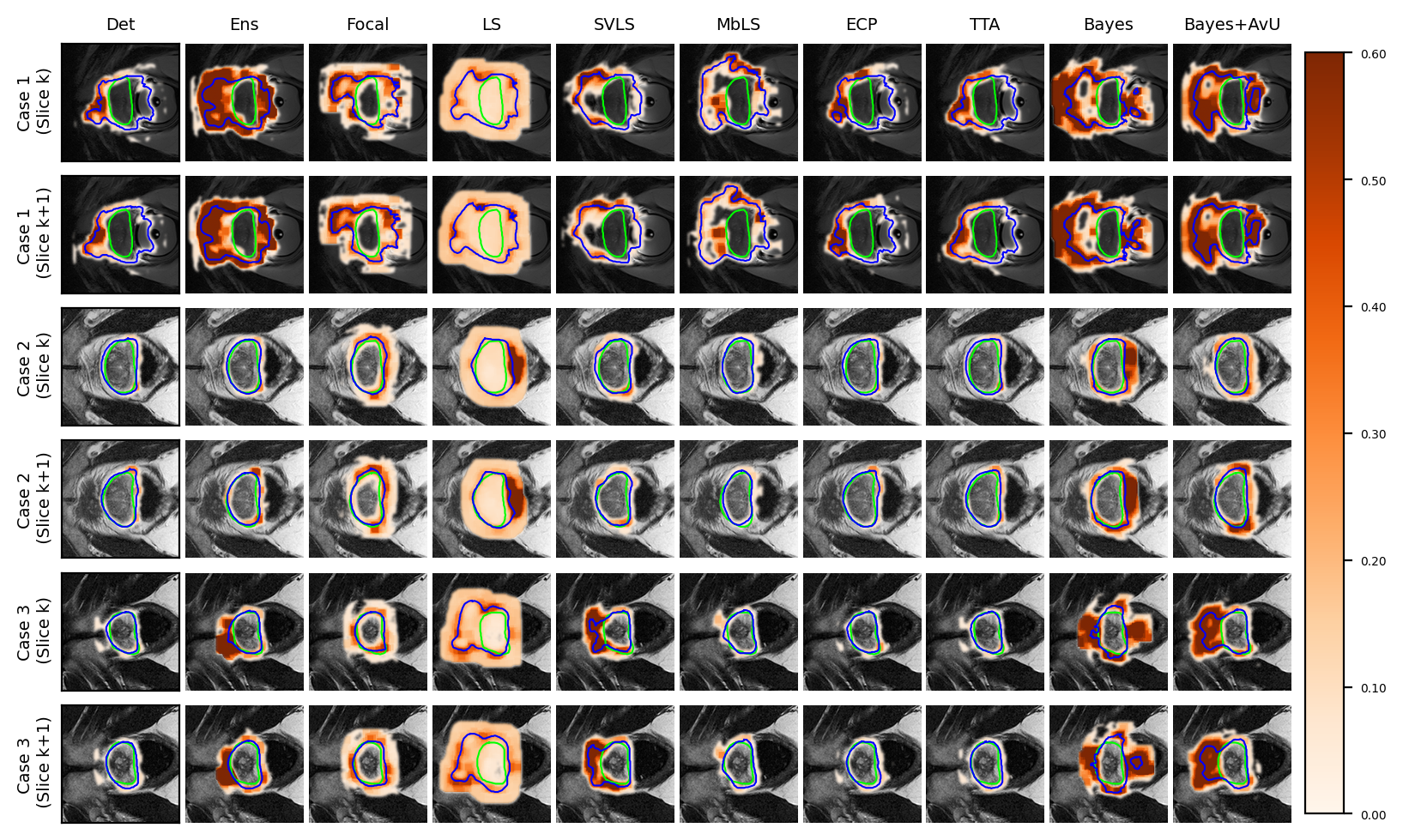
Visual results show that the Bayes+AvU successfully suppresses uncertainty in the true negative (Case 1 in Figure 3a, Case 2 in Figure 3b) and true positive (Case 2 in Figure 3a) regions of the predicted contour. It also shows uncertainty in the false positive regions (Case 2 in Figure 3a, Case 1/3 in Figure 3b)
5 Discussion
Although medical image segmentation using deep learning can now predict high quality contours which can be considered clinically acceptable, a manual quality assessment (QA) step is still required in a clinical setting. To truly make these models an integral part of clinical workflows, we need them to be able to express their uncertainty and for those uncertainties to be useful in a QA setting. To this end, we test 11 models which are either Bayesian, deterministic, calibration-focused or ensembled.
5.1 Discriminative and Calibrative Performance
In context of DICE and ECE, the use of the AvU loss on the baseline Bayes model always showed results which have never statistically deteriorated. Moreover, the DICE results for the in-distribution (ID) head-and-neck dataset (RTOG) were on-par with existing state-of-the-art models (83.6 vs 84.7 for Nikolov et al. (2021)). The same held for the ID Prostate dataset (PRMedDec) where results were better than advanced models (84.9 vs 83.0 for Antonelli et al. (2022)). These results validate the use of our neural architecture (Chen et al., 2021), and training strategy.
Secondly, although the Ensemble model, in general, had better or equivalent DICE and ECE scores across all 4 datasets, it also required 3x more parameters than the Bayes+AvU model. Also, as expected, and due to 5x more parameters, the Ensemble model performed better than the Det model for DICE and ECE.
Finally, in the regime of segmentation “failures” as the inaccuracy map, the calibrative methods did not generally have improved calibration performance when compared to the Det model. In theory, these models regularize the model’s probabilities by making it more uncertain and hence avoid overconfidence. In practice however, this leads to the predicted contours being uncertain along their accurate boundaries, most evident in visual examples of the Focal and SVLS model (see Figure 2 and Figure 3). Also, visual image characteristics in different regions of the scan that are similar to the segmented organs may cause these models to showcase uncertainty in those areas (for e.g. patches of uncertainty in Case 3 of Figure 2a).
5.2 Uncertainty-Error Correspondence Performance
Although calibrative metrics are useful to compare the average truthfulness of a model’s probabilities, they may not be relevant to real-world usage in a pixel-wise segmentation QA scenario. Considering a clinical workflow in which uncertainty can be used as a proxy for error-detection, we evaluate the correspondence between them. Results showed that across both in- and out-of-distribution datasets, the Bayes+AvU model has one of the highest uncertainty-error correspondence metrics. Similar trends were observed for the BayesH+AvU (Appendix E) model, however Bayes+AvU was better. We hypothesize that this is due to perturbations in the bottleneck of UNet-like models having a better understanding of semantic concepts (e.g., shape, size etc) than the decoder layers. However, the AvU loss did not offer benefit to the Det model on both datasets indicating that this loss may rely on the model to already exhibit some level of uncertainty.
An interesting case is shown in Figure 2b (Case 3) which showed uncertainty on the white blob (a vein) in the middle of the grey tissue of the organ. Many models showed uncertainty on the vein due to a difference in its texture from that of the organ. However, this information may be distracting to a clinician as they are using uncertainty for error detection. Given that there were no segmentation “failures”, our Bayes+AvU model successfully suppressed all uncertainties. In another case (Figure 2a - Case 3), we saw that for 3D segmentation, uncertainty is also 3D in nature. Our Bayes+AvU model had an error in the second slice and correctly showed uncertainty there. However, this uncertainty overflowed on the first slice and hence penalized the uncertainty-error correspondence metrics. Such results indicate that during contour QA, the clinician can potentially trust our AvU loss models more than other models as they are better indicative of potential errors. This reduces time wasted analyzing false positive regions (i.e., accurate but uncertain) and hence increases trust between an expert and deep learning-based contour QA tools. Also note that in general, the two-class prostate dataset visually showcased higher levels of uncertainty than the six-class head-and-neck dataset.
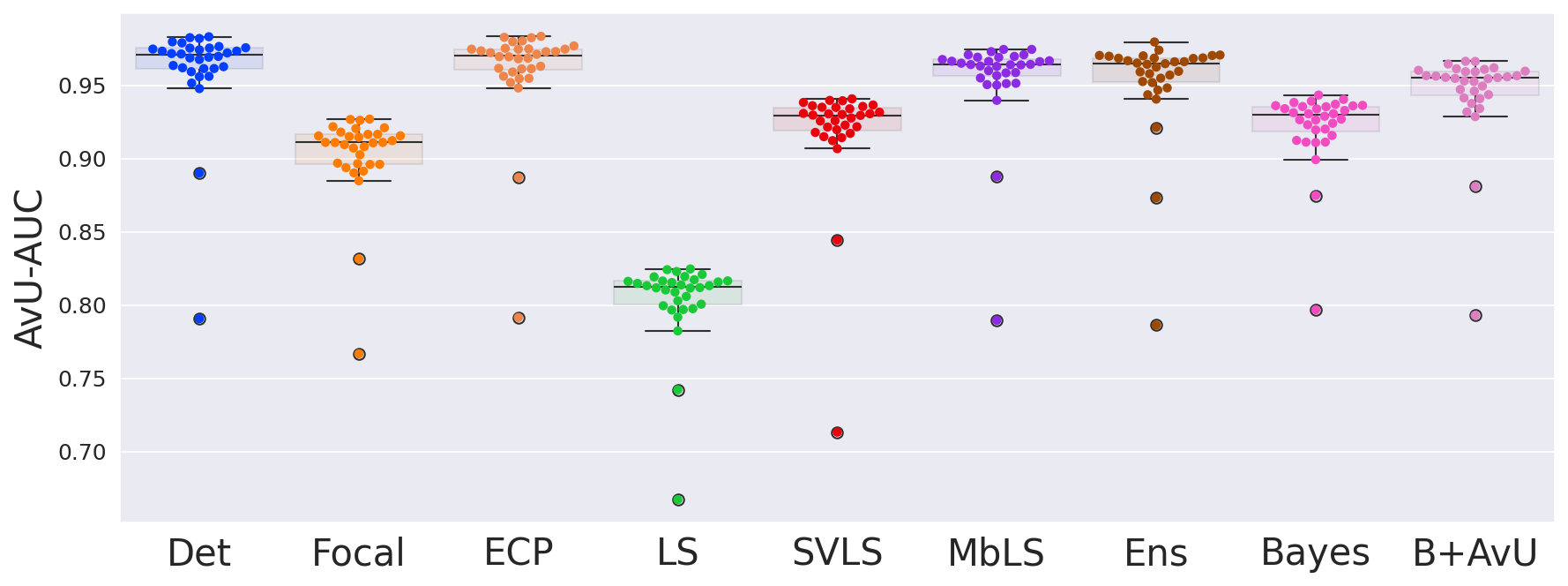
As seen in Table 1, Table 2 and Figure 4, there is no clear choice between the top two performing models i.e., Bayes+AvU and Ensemble for uncertainty-error correspondence. The visual results, however, indicate that the Ensemble model is more uncertain in accurate regions. Also, for all the datasets, the Det model has high AvU scores when compared to the Bayes+AvU model (Appendix C). Here, it is important to consider that the AvU metric (Equation 7) is essentially uncertainty accuracy, and thus, also comes with its own pitfalls. Given that all models had a DICE value which leads to more accurate terms and less inaccurate terms, the AvU metric got skewed due to the large count of terms. However, upon factoring the ROC and PRC curves, it becomes evident that the Det model is not the best performing for uncertainty-error correspondence.
Finally, all calibration-focused methods - Focal, ECP, LS, SVLS and MBLS had ROC and PRC metrics lower than the baseline Bayes model indicating that training for model calibration may not necessarily translate to uncertainty outputs useful for error detection.
5.3 Future Work
In a radiotherapy setting, the goal is to maximize radiation to tumorous regions and minimize it for healthy organs. This goal is often not optimally achieved due to imperfect contours caused by time constraints and amorphous region-of-interest boundaries on medical scans. Thus, an extension of our work could evaluate the contouring corrections made by clinicians in response to uncertainty-proposed errors in context of the dose changes to the different regions of interest. Such an experiment can better evaluate the clinical utility of an uncertainty-driven error correction workflow.
6 Conclusion
This work investigates the usage of the Accuracy-vs-Uncertainty (AvU) metric to improve clinical “utility” of deep Bayesian uncertainty as a proxy for error detection in segmentation settings. Experimental results indicate that using a differentiable AvU metric as an objective to train Bayesian segmentation models has a positive effect on uncertainty-error correspondence metrics. We show that our AvU-trained Bayesian models have equivalent or improved uncertainty-error correspondence metrics when compared to various calibrative and uncertainty-based methods. Given that our approach is a loss function, it can be used with other neural architectures capable of estimating uncertainty.
Given that deep learning models have shown the capability of reaching near expert-level performance in medical image segmentation, one of the next steps in their evolution is evaluating their clinical utility. Our work shows progress on this using a uncertainty-driven loss in a Bayesian setting. We do this for two radiotherapy body-sites and modalities as well in an out-of-distribution setting. Our hope is that the community is inspired by our positive results to further contribute to human-centric approaches to deep learning-based modeling.
Acknowledgments
The research for this work was funded by Varian, a Siemens Healthineers Company, through the HollandPTC-Varian Consortium (grant id 2019022) and partly financed by the Surcharge for Top Consortia for Knowledge and Innovation (TKIs) from the Ministry of Economic Affairs and Climate, The Netherlands.
Ethical Standards
The work follows appropriate ethical standards in conducting research and writing the manuscript, following all applicable laws and regulations regarding treatment of animals or human subjects.
Conflicts of Interest
We declare we don’t have conflicts of interest.
References
- Ang et al. (2014) K Kian Ang, Qiang Zhang, David I Rosenthal, Phuc Felix Nguyen-Tan, Eric J Sherman, Randal S Weber, James M Galvin, James A Bonner, Jonathan Harris, Adel K El-Naggar, et al. Randomized phase iii trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage iii to iv head and neck carcinoma: Rtog 0522. Journal of clinical oncology, 32(27):2940, 2014.
- Antonelli et al. (2022) Michela Antonelli, Annika Reinke, Spyridon Bakas, Keyvan Farahani, Annette Kopp-Schneider, Bennett A Landman, Geert Litjens, Bjoern Menze, Olaf Ronneberger, Ronald M Summers, et al. The medical segmentation decathlon. Nature communications, 13(1):4128, 2022.
- Bhat et al. (2022) Ishaan Bhat, Josien P.W. Pluim, Max A. Viergever, and Hugo J. Kuijf. Influence of uncertainty estimation techniques on false-positive reduction in liver lesion detection. Machine Learning for Biomedical Imaging, 1:1–33, 2022. ISSN 2766-905X. . URL https://melba-journal.org/2022:030.
- Blundell et al. (2015) Charles Blundell, Julien Cornebise, Koray Kavukcuoglu, and Daan Wierstra. Weight uncertainty in neural network. In International conference on machine learning, pages 1613–1622. PMLR, 2015.
- Bragman et al. (2018) Felix JS Bragman, Ryutaro Tanno, Zach Eaton-Rosen, Wenqi Li, David J Hawkes, Sebastien Ourselin, Daniel C Alexander, Jamie R McClelland, and M Jorge Cardoso. Uncertainty in multitask learning: joint representations for probabilistic mr-only radiotherapy planning. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part IV 11, pages 3–11. Springer, 2018.
- Brouwer et al. (2012) Charlotte L Brouwer, Roel JHM Steenbakkers, Edwin van den Heuvel, Joop C Duppen, Arash Navran, Henk P Bijl, Olga Chouvalova, Fred R Burlage, Harm Meertens, Johannes A Langendijk, et al. 3D variation in delineation of head and neck organs at risk. Radiation Oncology, 7(1):1–10, 2012.
- Brouwer et al. (2020) Charlotte L Brouwer, Djamal Boukerroui, Jorge Oliveira, Padraig Looney, Roel JHM Steenbakkers, Johannes A Langendijk, Stefan Both, and Mark J Gooding. Assessment of manual adjustment performed in clinical practice following deep learning contouring for head and neck organs at risk in radiotherapy. Physics and imaging in radiation oncology, 16:54–60, 2020.
- Camarasa et al. (2021) Robin Camarasa, Daniel Bos, Jeroen Hendrikse, Paul Nederkoorn, M Eline Kooi, Aad van der Lugt, Marleen de Bruijne, et al. A quantitative comparison of epistemic uncertainty maps applied to multi-class segmentation. Machine Learning for Biomedical Imaging, 1(UNSURE2020 special issue):1–10, 2021.
- Chaves-de-Plaza et al. (2022) Nicolas F Chaves-de-Plaza, Prerak Mody, Klaus Hildebrandt, Marius Staring, Eleftheria Astreinidou, Mischa de Ridder, Huib de Ridder, and René van Egmond. Towards fast human-centred contouring workflows for adaptive external beam radiotherapy. In Proceedings of the Human Factors and Ergonomics Society Europe Chapter 2022 Annual Conference, 2022.
- Chen et al. (2021) Zijie Chen, Cheng Li, Junjun He, Jin Ye, Diping Song, Shanshan Wang, Lixu Gu, and Yu Qiao. A novel hybrid convolutional neural network for accurate organ segmentation in 3D head and neck CT images. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 569–578. Springer, 2021.
- Dawid (1982) A Philip Dawid. The well-calibrated Bayesian. Journal of the American Statistical Association, 77(379):605–610, 1982.
- Diaz-Pinto et al. (2022) Andres Diaz-Pinto, Pritesh Mehta, Sachidanand Alle, Muhammad Asad, Richard Brown, Vishwesh Nath, Alvin Ihsani, Michela Antonelli, Daniel Palkovics, Csaba Pinter, et al. Deepedit: Deep editable learning for interactive segmentation of 3D medical images. In Data Augmentation, Labelling, and Imperfections: Second MICCAI Workshop, DALI 2022, Held in Conjunction with MICCAI 2022, pages 11–21. Springer, 2022.
- Ding et al. (2021) Zhipeng Ding, Xu Han, Peirong Liu, and Marc Niethammer. Local temperature scaling for probability calibration. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 6889–6899, 2021.
- Gal (2016) Yarin Gal. Uncertainty in deep learning. PhD Thesis, University of Cambridge, 2016.
- Gal and Ghahramani (2016) Yarin Gal and Zoubin Ghahramani. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In International conference on machine learning, pages 1050–1059. PMLR, 2016.
- Garifullin et al. (2021) Azat Garifullin, Lasse Lensu, and Hannu Uusitalo. Deep Bayesian baseline for segmenting diabetic retinopathy lesions: Advances and challenges. Computers in Biology and Medicine, 136:104725, 2021.
- Ghoshal and Tucker (2022) Biraja Ghoshal and Allan Tucker. On calibrated model uncertainty in deep learning, 2022. URL https://europepmc.org/article/PPR/PPR517139.
- Gibson et al. (2022) Eli Gibson, Bogdan Georgescu, Pascal Ceccaldi, Pierre-Hugo Trigan, Youngjin Yoo, Jyotipriya Das, Thomas J Re, Vishwanath Rs, Abishek Balachandran, Eva Eibenberger, et al. Artificial intelligence with statistical confidence scores for detection of acute or subacute hemorrhage on noncontrast CT head scans. Radiology: Artificial Intelligence, 4(3):e210115, 2022.
- Grepl et al. (2023) Jakub Grepl, Igor Sirak, Milan Vosmik, Denisa Pohankova, Miroslav Hodek, Petr Paluska, and Ales Tichy. Mri-based adaptive radiotherapy has the potential to reduce dysphagia in patients with head and neck cancer. Physica Medica, 105:102511, 2023.
- Gruber and Buettner (2022) Sebastian Gruber and Florian Buettner. Better uncertainty calibration via proper scores for classification and beyond. Advances in Neural Information Processing Systems, 35:8618–8632, 2022.
- Guo et al. (2017) Chuan Guo, Geoff Pleiss, Yu Sun, and Kilian Q Weinberger. On calibration of modern neural networks. In International conference on machine learning, pages 1321–1330. PMLR, 2017.
- Hekler et al. (2023) Achim Hekler, Titus J Brinker, and Florian Buettner. Test time augmentation meets post-hoc calibration: uncertainty quantification under real-world conditions. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 37, pages 14856–14864, 2023.
- Hu et al. (2019) Shi Hu, Daniel Worrall, Stefan Knegt, Bas Veeling, Henkjan Huisman, and Max Welling. Supervised uncertainty quantification for segmentation with multiple annotations. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part II 22, pages 137–145. Springer, 2019.
- Islam and Glocker (2021) Mobarakol Islam and Ben Glocker. Spatially varying label smoothing: Capturing uncertainty from expert annotations. In Information Processing in Medical Imaging: 27th International Conference, IPMI 2021, Virtual Event, June 28–June 30, 2021, Proceedings 27, pages 677–688. Springer, 2021.
- Iwamoto et al. (2021) Sora Iwamoto, Bisser Raytchev, Toru Tamaki, and Kazufumi Kaneda. Improving the reliability of semantic segmentation of medical images by uncertainty modeling with Bayesian deep networks and curriculum learning. In Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis. Springer, 2021.
- Karimi and Gholipour (2022) Davood Karimi and Ali Gholipour. Improving calibration and out-of-distribution detection in deep models for medical image segmentation. IEEE Transactions on Artificial Intelligence, 4(2):383–397, 2022.
- Karimi et al. (2019) Davood Karimi, Qi Zeng, Prateek Mathur, Apeksha Avinash, Sara Mahdavi, Ingrid Spadinger, Purang Abolmaesumi, and Septimiu E Salcudean. Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images. Medical image analysis, 57:186–196, 2019.
- Krishnan and Tickoo (2020) Ranganath Krishnan and Omesh Tickoo. Improving model calibration with accuracy versus uncertainty optimization. Advances in Neural Information Processing Systems, 2020.
- Kumar et al. (2019) Ananya Kumar, Percy S Liang, and Tengyu Ma. Verified uncertainty calibration. Advances in Neural Information Processing Systems, 32, 2019.
- Laves et al. (2019) Max-Heinrich Laves, Sontje Ihler, Karl-Philipp Kortmann, and Tobias Ortmaier. Well-calibrated model uncertainty with temperature scaling for dropout variational inference. arXiv preprint arXiv:1909.13550, 2019.
- Lei et al. (2019) Wenhui Lei, Huan Wang, Ran Gu, Shichuan Zhang, Shaoting Zhang, and Guotai Wang. Deepigeos-v2: deep interactive segmentation of multiple organs from head and neck images with lightweight cnns. In Large-Scale Annotation of Biomedical Data and Expert Label Synthesis and Hardware Aware Learning for Medical Imaging and Computer Assisted Intervention: International Workshops, LABELS 2019, HAL-MICCAI 2019, and CuRIOUS 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, Proceedings 4, pages 61–69. Springer, 2019.
- Lin et al. (2017) Tsung-Yi Lin, Priya Goyal, Ross Girshick, Kaiming He, and Piotr Dollár. Focal loss for dense object detection. In Proceedings of the IEEE international conference on computer vision, pages 2980–2988, 2017.
- Litjens et al. (2014) Geert Litjens, Robert Toth, Wendy van de Ven, Caroline Hoeks, Sjoerd Kerkstra, Bram van Ginneken, Graham Vincent, Gwenael Guillard, Neil Birbeck, Jindang Zhang, et al. Evaluation of prostate segmentation algorithms for mri: the promise12 challenge. Medical image analysis, 18(2):359–373, 2014.
- Liu et al. (2022) Bingyuan Liu, Ismail Ben Ayed, Adrian Galdran, and Jose Dolz. The devil is in the margin: Margin-based label smoothing for network calibration. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 80–88, 2022.
- Liu et al. (2023) Bingyuan Liu, Jérôme Rony, Adrian Galdran, Jose Dolz, and Ismail Ben Ayed. Class adaptive network calibration. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16070–16079, 2023.
- Mehrtash et al. (2020) Alireza Mehrtash, William M Wells, Clare M Tempany, Purang Abolmaesumi, and Tina Kapur. Confidence calibration and predictive uncertainty estimation for deep medical image segmentation. IEEE transactions on medical imaging, 39(12):3868–3878, 2020.
- Meyer et al. (2021) Anneke Meyer, Grzegorz Chlebus, Marko Rak, Daniel Schindele, Martin Schostak, Bram van Ginneken, Andrea Schenk, Hans Meine, Horst K Hahn, Andreas Schreiber, et al. Anisotropic 3D multi-stream cnn for accurate prostate segmentation from multi-planar mri. Computer Methods and Programs in Biomedicine, 200:105821, 2021.
- Mody et al. (2022a) Prerak Mody, Nicolas F. Chaves-de-Plaza, Klaus Hildebrandt, and Marius Staring. Improving Error Detection in Deep Learning Based Radiotherapy Autocontouring Using Bayesian Uncertainty. In Uncertainty for Safe Utilization of Machine Learning in Medical Imaging: 4th International Workshop, UNSURE 2022, Held in Conjunction with MICCAI 2022, 2022a. ISBN 978-3-031-16748-5.
- Mody et al. (2022b) Prerak P Mody, Nicolas Chaves-de-Plaza, Klaus Hildebrandt, René van Egmond, Huib de Ridder, and Marius Staring. Comparing bayesian models for organ contouring in head and neck radiotherapy. In Medical Imaging 2022: Image Processing, volume 12032, pages 100–109. SPIE, 2022b.
- Monteiro et al. (2020) Miguel Monteiro, Loïc Le Folgoc, Daniel Coelho de Castro, Nick Pawlowski, Bernardo Marques, Konstantinos Kamnitsas, Mark van der Wilk, and Ben Glocker. Stochastic segmentation networks: Modelling spatially correlated aleatoric uncertainty. Advances in Neural Information Processing Systems, 33:12756–12767, 2020.
- Mukhoti and Gal (2018) Jishnu Mukhoti and Yarin Gal. Evaluating Bayesian deep learning methods for semantic segmentation. CoRR, abs/1811.12709, 2018.
- Mukhoti et al. (2020) Jishnu Mukhoti, Viveka Kulharia, Amartya Sanyal, Stuart Golodetz, Philip Torr, and Puneet Dokania. Calibrating deep neural networks using focal loss. Advances in Neural Information Processing Systems, 33:15288–15299, 2020.
- Müller et al. (2019) Rafael Müller, Simon Kornblith, and Geoffrey E Hinton. When does label smoothing help? Advances in neural information processing systems, 32, 2019.
- Murugesan et al. (2023a) Balamurali Murugesan, Sukesh Adiga Vasudeva, Bingyuan Liu, Hervé Lombaert, Ismail Ben Ayed, and Jose Dolz. Trust your neighbours: Penalty-based constraints for model calibration. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 572–581. Springer, 2023a.
- Murugesan et al. (2023b) Balamurali Murugesan, Bingyuan Liu, Adrian Galdran, Ismail Ben Ayed, and Jose Dolz. Calibrating segmentation networks with margin-based label smoothing. Medical Image Analysis, 87:102826, 2023b.
- Nair et al. (2020) Tanya Nair, Doina Precup, Douglas L Arnold, and Tal Arbel. Exploring uncertainty measures in deep networks for multiple sclerosis lesion detection and segmentation. Medical image analysis, 59:101557, 2020.
- Ng et al. (2022) Matthew Ng, Fumin Guo, Labonny Biswas, Steffen E. Petersen, Stefan K. Piechnik, Stefan Neubauer, and Graham Wright. Estimating uncertainty in neural networks for cardiac MRI segmentation: A benchmark study. IEEE Transactions on Biomedical Engineering, pages 1–12, 2022. .
- Nikolov et al. (2021) Stanislav Nikolov, Sam Blackwell, Alexei Zverovitch, Ruheena Mendes, Michelle Livne, Jeffrey De Fauw, Yojan Patel, Clemens Meyer, Harry Askham, Bernadino Romera-Paredes, et al. Clinically applicable segmentation of head and neck anatomy for radiotherapy: deep learning algorithm development and validation study. Journal of Medical Internet Research, 23(7):e26151, 2021.
- Nixon et al. (2019) Jeremy Nixon, Michael W Dusenberry, Linchuan Zhang, Ghassen Jerfel, and Dustin Tran. Measuring calibration in deep learning. In CVPR workshops, volume 2, 2019.
- Ouyang et al. (2022) Cheng Ouyang, Shuo Wang, Chen Chen, Zeju Li, Wenjia Bai, Bernhard Kainz, and Daniel Rueckert. Improved post-hoc probability calibration for out-of-domain MRI segmentation. In Uncertainty for Safe Utilization of Machine Learning in Medical Imaging: 4th International Workshop, UNSURE 2022, Held in Conjunction with MICCAI 2022, pages 59–69. Springer, 2022.
- Ovadia et al. (2019) Yaniv Ovadia, Emily Fertig, Jie Ren, Zachary Nado, David Sculley, Sebastian Nowozin, Joshua Dillon, Balaji Lakshminarayanan, and Jasper Snoek. Can you trust your model’s uncertainty? evaluating predictive uncertainty under dataset shift. Advances in neural information processing systems, 32, 2019.
- Pereyra et al. (2017) Gabriel Pereyra, George Tucker, Jan Chorowski, Łukasz Kaiser, and Geoffrey Hinton. Regularizing neural networks by penalizing confident output distributions. arXiv preprint arXiv:1701.06548, 2017.
- Petragallo et al. (2022) Rachel Petragallo, Naomi Bardach, Ezequiel Ramirez, and James M Lamb. Barriers and facilitators to clinical implementation of radiotherapy treatment planning automation: A survey study of medical dosimetrists. Journal of Applied Clinical Medical Physics, 23(5):e13568, 2022.
- Raudaschl et al. (2017) Patrik F Raudaschl, Paolo Zaffino, Gregory C Sharp, Maria Francesca Spadea, Antong Chen, Benoit M Dawant, Thomas Albrecht, Tobias Gass, Christoph Langguth, Marcel Lüthi, et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Medical physics, 44(5):2020–2036, 2017.
- Sambaturu et al. (2023) Bhavani Sambaturu, Ashutosh Gupta, CV Jawahar, and Chetan Arora. Scribblenet: Efficient interactive annotation of urban city scenes for semantic segmentation. Pattern Recognition, 133:109011, 2023.
- Sander et al. (2020) Jörg Sander, Bob D de Vos, and Ivana Išgum. Automatic segmentation with detection of local segmentation failures in cardiac mri. Scientific Reports, 10(1):21769, 2020.
- Soberanis-Mukul et al. (2020) Roger D Soberanis-Mukul, Nassir Navab, and Shadi Albarqouni. Uncertainty-based graph convolutional networks for organ segmentation refinement. In Medical Imaging with Deep Learning, pages 755–769. PMLR, 2020.
- Szegedy et al. (2016) Christian Szegedy, Vincent Vanhoucke, Sergey Ioffe, Jon Shlens, and Zbigniew Wojna. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 2818–2826, 2016.
- Taghanaki et al. (2019) Saeid Asgari Taghanaki, Yefeng Zheng, S Kevin Zhou, Bogdan Georgescu, Puneet Sharma, Daguang Xu, Dorin Comaniciu, and Ghassan Hamarneh. Combo loss: Handling input and output imbalance in multi-organ segmentation. Computerized Medical Imaging and Graphics, 75:24–33, 2019.
- Thulasidasan et al. (2019) Sunil Thulasidasan, Gopinath Chennupati, Jeff A Bilmes, Tanmoy Bhattacharya, and Sarah Michalak. On mixup training: Improved calibration and predictive uncertainty for deep neural networks. Advances in neural information processing systems, 32, 2019.
- van der Veen et al. (2019) Julie van der Veen, Akos Gulyban, and Sandra Nuyts. Interobserver variability in delineation of target volumes in head and neck cancer. Radiotherapy and Oncology, 137:9–15, 2019.
- Van Dijk et al. (2020) Lisanne V Van Dijk, Lisa Van den Bosch, Paul Aljabar, Devis Peressutti, Stefan Both, Roel JHM Steenbakkers, Johannes A Langendijk, Mark J Gooding, and Charlotte L Brouwer. Improving automatic delineation for head and neck organs at risk by deep learning contouring. Radiotherapy and Oncology, 142:115–123, 2020.
- Wan et al. (2013) Li Wan, Matthew Zeiler, Sixin Zhang, Yann Le Cun, and Rob Fergus. Regularization of neural networks using dropconnect. In International conference on machine learning, pages 1058–1066. PMLR, 2013.
- Wang et al. (2019a) Guotai Wang, Wenqi Li, Michael Aertsen, Jan Deprest, Sébastien Ourselin, and Tom Vercauteren. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing, 338:34–45, 2019a.
- Wang et al. (2019b) Guotai Wang, Wenqi Li, Michael Aertsen, Jan Deprest, Sébastien Ourselin, and Tom Vercauteren. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing, 338:34–45, 2019b.
- Wang et al. (2020) Guotai Wang, Michael Aertsen, Jan Deprest, Sébastien Ourselin, Tom Vercauteren, and Shaoting Zhang. Uncertainty-guided efficient interactive refinement of fetal brain segmentation from stacks of mri slices. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part IV 23, pages 279–288. Springer, 2020.
- Wen et al. (2018) Yeming Wen, Paul Vicol, Jimmy Ba, Dustin Tran, and Roger Grosse. Flipout: Efficient pseudo- independent weight perturbations on mini-batches. In Proceedings of the 6th International Conference on Learning Representations, 2018.
- Ye et al. (2022) Xianghua Ye, Dazhou Guo, Jia Ge, Senxiang Yan, Yi Xin, Yuchen Song, Yongheng Yan, Bing-shen Huang, Tsung-Min Hung, Zhuotun Zhu, et al. Comprehensive and clinically accurate head and neck cancer organs-at-risk delineation on a multi-institutional study. Nature Communications, 13(1):6137, 2022.
- Yeung et al. (2022) Michael Yeung, Evis Sala, Carola-Bibiane Schönlieb, and Leonardo Rundo. Unified focal loss: Generalising dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Computerized Medical Imaging and Graphics, 95:102026, 2022.
- Zabel et al. (2021) W Jeffrey Zabel, Jessica L Conway, Adam Gladwish, Julia Skliarenko, Giulio Didiodato, Leah Goorts-Matthews, Adam Michalak, Sarah Reistetter, Jenna King, Keith Nakonechny, et al. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Practical Radiation Oncology, 11(1):e80–e89, 2021.
- Zhang et al. (2020) Jize Zhang, Bhavya Kailkhura, and T Yong-Jin Han. Mix-n-match: Ensemble and compositional methods for uncertainty calibration in deep learning. In International conference on machine learning, pages 11117–11128. PMLR, 2020.
Appendix A Segmentation “Failures” and “Errors”
false



Appendix B Weightage of AvU loss
The table below show the weights used for the AvU loss which were finetuned on the validation datasets of the head-and-neck CT and prostate MR. The final weightage was chosen by identifying the inflection point at which the ROC-AUC and PRC-AUC drop precipitously. Given that the AvU loss is a log term, its values are inherently small ( 1.0). This is then added to the cross-entropy term, which is a sum of logs (Eqn (3)) over all the voxels (=N) and all the classes (=C). Thus, we used a balancing term in the range of 101 to 104.
| ValidationDataset | Model | AvU-AUC(x) | ROC-AUC(x) | PRC-AUC(x) |
|---|---|---|---|---|
| H&N CT(MICCAI2015) | Bayes | 34.1 ± 0.7 | 79.1 ± 4.7 | 25.9 ± 2.9 |
| Bayes + 10AvU | 34.5 ± 0.9 | 78.2 ± 6.0 | 26.1 ± 3.4 | |
| Bayes + 100AvU | 35.5 ± 0.6 | 79.6 ± 4.8 | 28.0 ± 3.5 | |
| Bayes + 1000AvU | 35.9 ± 6.9 | 76.4 ± 5.8 | 23.1 ± 1.7 | |
| ProstateMR(ProstateX) | Bayes | 93.2 ± 1.8 | 95.3 ± 1.9 | 30.3 ± 2.9 |
| Bayes + 100AvU | 94.9 ± 2.1 | 95.9 ± 2.0 | 31.5 ± 3.5 | |
| Bayes + 1000AvU | 95.5 ± 1.9 | 96.3 ± 2.4 | 32.0 ± 3.3 | |
| Bayes + 10000AvU | 96.1 ± 1.7 | 93.1 ± 2.1 | 29.3 ± 3.1 |
Appendix C Hyperparameter selection
In the tables shown below, we report results for different hyperparameters of different model classes. If the DICE of a hyperparameter is 10.0 points lower than the class maximum, we ignore it. We also ignore models with large drops in ECE or AvU-AUC when compared to models in its own class. To choose the best hyperparameter, it has to perform as the best in four out of the five metrics, else we chose the middlemost hyperparameter.
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 83.6 ± 2.2 | 8.5 ± 1.6 | 35.1 ± 1.1 | 74.8 ± 5.0 | 24.5 ± 0.9 |
| Det + 10AvU | 83.4 ± 1.7 | 8.4 ± 1.6 | 35.4 ± 1.0 | 74.4 ± 4.8 | 23.2 ± 1.4 |
| Det + 100AvU | 83.6 ± 1.4 | 8.1 ± 1.5 | 36.1 ± 0.6 | 75.6 ± 2.9 | 23.4 ± 2.1 |
| Det + 1000AvU | 58.1 ± 6.9 | 14.7 ± 4.0 | 30.2 ± 1.6 | 78.8 ± 4.6 | 23.0 ± 10.6 |
| 1-6 Focal(=1) | 84.1 ± 0.8 | 8.0 ± 0.6 | 32.4 ± 0.7 | 73.9 ± 4.1 | 21.5 ± 3.4 |
| Focal(=2) | 83.4 ± 1.3 | 9.6 ± 8.3 | 24.8 ± 1.1 | 73.7 ± 1.6 | 22.7 ± 4.1 |
| Focal(=3) | 84.1 ± 1.9 | 15.5 ± 1.9 | 17.5 ± 7.4 | 73.1 ± 3.2 | 12.6 ± 1.9 |
| 1-6 ECP(=0.1) | 83.9 ± 1.3 | 8.3 ± 1.3 | 35.3 ± 0.8 | 75.1 ± 4.3 | 22.3 ± 0.8 |
| ECP(=1.0) | 84.0 ± 1.1 | 8.5 ± 0.8 | 35.4 ± 0.7 | 75.3 ± 3.2 | 23.4 ± 1.4 |
| ECP(=10.0) | 83.2 ± 2.1 | 8.7 ± 1.3 | 35.2 ± 0.8 | 74.9 ± 3.9 | 24.6 ± 2.1 |
| ECP(=100.0) | 81.2 ± 6.4 | 17.9 ± 1.5 | 28.7 ± 2.8 | 65.4 ± 5.6 | 17.5 ± 5.2 |
| 1-6 LS(=0.01) | 83.0 ± 2.1 | 8.1 ± 1.3 | 32.6 ± 0.9 | 70.9 ± 2.6 | 23.4 ± 2.7 |
| LS(=0.05) | 83.6 ± 1.2 | 6.1 ± 1.0 | 24.9 ± 0.4 | 64.5 ± 3.3 | 18.1 ± 2.0 |
| LS(=0.1) | 83.5 ± 1.2 | 7.9 ± 1.2 | 17.5 ± 0.1 | 63.9 ± 2.2 | 22.2 ± 1.1 |
| 1-6 SVLS(=1) | 83.5 ± 1.3 | 7.7 ± 0.7 | 32.3 ± 0.8 | 71.5 ± 2.5 | 19.9 ± 0.4 |
| SVLS(=2) | 83.5 ± 1.7 | 8.1 ± 0.9 | 31.8 ± 1.0 | 70.5 ± 3.8 | 17.7 ± 1.5 |
| SVLS(=3) | 84.1 ± 2.0 | 7.7 ± 0.7 | 31.9 ± 1.0 | 71.3 ± 4.4 | 19.2 ± 3.2 |
| 1-6 MbLS(,m=30) | 82.7 ± 1.8 | 8.5 ± 0.6 | 34.9 ± 1.0 | 74.0 ± 4.3 | 23.1 ± 1.2 |
| MbLS(,m=20) | 84.4 ± 1.4 | 8.0 ± 1.1 | 35.2 ± 0.7 | 72.3 ± 3.3 | 20.4 ± 1.0 |
| MbLS(,m=10) | 82.7 ± 1.8 | 8.5 ± 0.6 | 32.9 ± 0.7 | 68.4 ± 3.0 | 21.7 ± 2.2 |
| MbLS(,m=8) | 62.9 ± 7.6 | 18.75 ± 1.4 | 26.0 ± 0.4 | 74.9 ± 4.0 | 39.1 ± 2.7 |
| [0.5pt/5pt] MbLS(,m=20) | 83.2 ± 1.3 | 8.9 ± 1.5 | 35.0 ± 0.9 | 72.4 ± 4.4 | 22.5 ± 1.1 |
| MbLS(,m=20) | 83.4 ± 1.4 | 8.5 ± 2.0 | 34.2 ± 1.1 | 72.2 ± 4.4 | 23.1 ± 2.0 |
| MbLS(,m=20) | 81.8 ± 1.8 | 8.0 ± 1.1 | 32.1 ± 0.9 | 69.6 ± 4.8 | 21.0 ± 2.3 |
| 1-6 TTA | 83.5 ± 2.2 | 8.5 ± 1.7 | 34.9 ± 1.1 | 75.3 ± 5.2 | 25.2 ± 1.7 |
| 1-6 Ens | 84.9 ± 1.6 | 6.8 ± 0.9 | 34.1 ± 1.1 | 80.8 ± 3.2 | 28.2 ± 4.1 |
| 1-6 Bayes | 84.2 ± 2.9 | 7.8 ± 1.3 | 34.1 ± 0.7 | 79.1 ± 4.7 | 25.9 ± 2.9 |
| Bayes + 10AvU | 83.1 ± 2.9 | 7.6 ± 2.0 | 34.5 ± 0.9 | 78.2 ± 6.0 | 26.1 ± 3.4 |
| Bayes + 100AvU | 83.2 ± 1.7 | 7.0 ± 1.9 | 35.5 ± 0.6 | 79.6 ± 4.8 | 28.0 ± 3.5 |
| Bayes + 1000AvU | 84.3 ± 1.0 | 7.5 ± 1.5 | 35.9 ± 6.9 | 76.4 ± 5.8 | 23.1 ± 1.7 |
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 84.2 ± 2.7 | 9.0 ± 2.1 | 35.5 ± 1.5 | 73.0 ± 5.7 | 21.0 ± 4.8 |
| Det + 10AvU | 83.7 ± 2.3 | 9.3 ± 2.2 | 35.7 ± 1.3 | 70.6 ± 5.3 | 20.0 ± 3.6 |
| Det + 100AvU* | 83.8 ± 2.9 | 8.6 ± 2.7 | 36.2 ± 1.4 | 73.1 ± 6.0 | 20.8 ± 4.0 |
| Det + 1000AvU | 62.3 ± 5.6 | 12.1 ± 2.9 | 30.7 ± 1.2 | 78.0 ± 4.6 | 16.0 ± 9.0 |
| 1-6 Focal(=1)* | 84.3 ± 2.4 | 9.3 ± 1.5 | 32.5 ± 0.9 | 70.3 ± 5.5 | 18.2 ± 3.2 |
| Focal(=2) | 84.2 ± 2.0 | 11.2 ± 1.6 | 25.1 ± 0.7 | 69.4 ± 4.9 | 17.2 ± 3.0 |
| Focal(=3) | 83.9 ± 2.5 | 15.7 ± 2.3 | 17.9 ± 5.3 | 70.5 ± 5.0 | 12.2 ± 2.9 |
| 1-6 ECP(=0.1) | 84.4 ± 2.2 | 8.9 ± 2.1 | 35.7 ± 1.3 | 72.9 ± 6.3 | 20.1 ± 3.8 |
| ECP(=1.0)* | 84.4 ± 2.3 | 9.0 ± 2.0 | 35.9 ± 1.3 | 73.8 ± 5.4 | 21.0 ± 3.7 |
| ECP(=10.0) | 84.3 ± 2.7 | 9.2 ± 2.4 | 35.8 ± 1.4 | 73.5 ± 6.0 | 20.6 ± 4.3 |
| ECP(=100.0) | 70.8 ± 3.9 | 18.6 ± 2.8 | 21.4 ± 2.7 | 58.7 ± 2.6 | 28.9 ± 7.1 |
| 1-6 LS(=0.01) | 83.4 ± 2.8 | 9.0 ± 2.9 | 32.9 ± 0.1 | 66.1 ± 5.7 | 18.4 ± 3.6 |
| LS(=0.05)* | 83.0 ± 3.0 | 7.5 ± 2.2 | 25.1 ± 0.5 | 62.6 ± 3.3 | 17.5 ± 4.0 |
| LS(=0.1) | 84.1 ± 2.3 | 8.4 ± 2.9 | 17.5 ± 0.1 | 62.3 ± 2.5 | 18.5 ± 3.5 |
| 1-6 SVLS(=1)* | 83.9 ± 2.5 | 9.0 ± 2.3 | 32.6 ± 1.1 | 69.6 ± 8.3 | 18.8 ± 2.8 |
| SVLS(=2) | 84.2 ± 2.6 | 9.0 ± 2.0 | 32.2 ± 1.1 | 70.8 ± 7.1 | 18.1 ± 3.5 |
| SVLS(=3) | 83.9 ± 2.7 | 9.0 ± 2.2 | 32.1 ± 1.2 | 69.3 ± 7.0 | 18.8 ± 2.8 |
| 1-6 MbLS(,m=30) | 83.7 ± 2.6 | 9.0 ± 2.0 | 35.4 ± 1.2 | 70.0 ± 5.6 | 19.7 ± 4.1 |
| MbLS(,m=20)* | 84.0 ± 2.6 | 9.2 ± 2.1 | 35.3 ± 1.3 | 67.5 ± 5.7 | 19.5 ± 3.5 |
| MbLS(,m=10) | 82.4 ± 2.6 | 9.8 ± 2.8 | 33.1 ± 1.2 | 64.1 ± 7.0 | 18.3 ± 3.1 |
| MbLS(,m=8) | 62.4 ± 8.2 | 18.9 ± 1.6 | 26.3 ± 0.4 | 73.6 ± 6.6 | 38.3 ± 4.1 |
| [0.5pt/5pt] MbLS(,m=20) | 83.4 ± 2.5 | 9.2 ± 2.6 | 35.4 ± 1.3 | 71.3 ± 7.0 | 20.1 ± 3.6 |
| MbLS(,m=20) | 83.0 ± 3.4 | 9.5 ± 2.8 | 34.6 ± 1.4 | 69.1 ± 6.0 | 20.1 ± 4.1 |
| MbLS(,m=20) | 82.5 ± 3.2 | 9.1 ± 3.0 | 32.4 ± 1.4 | 68.2 ± 7.3 | 19.0 ± 2.9 |
| 1-6 TTA | 84.1 ± 2.8 | 9.1 ± 2.1 | 35.5 ± 1.4 | 72.9 ± 5.9 | 20.8 ± 3.9 |
| 1-6 Ens | 85.0 ± 2.7 | 7.8 ± 1.9 | 34.5 ± 1.2 | 78.6 ± 4.7 | 25.7 ± 6.8 |
| 1-6 Bayes | 83.9 ± 2.6 | 8.7 ± 2.1 | 34.5 ± 1.2 | 74.1 ± 5.4 | 22.1 ± 3.5 |
| Bayes + 10AvU | 83.4 ± 2.8 | 8.7 ± 2.4 | 34.7 ± 1.3 | 74.7 ± 4.9 | 24.4 ± 4.1 |
| Bayes + 100AvU* | 83.6 ± 2.5 | 7.6 ± 2.5 | 35.6 ± 1.2 | 76.1 ± 5.6 | 25.1 ± 5.3 |
| Bayes + 1000AvU | 83.5 ± 3.0 | 8.5 ± 3.4 | 36.1 ± 1.5 | 77.2 ± 6.0 | 24.7 ± 4.5 |
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 78.1 ± 4.6 | 12.9 ± 2.6 | 33.4 ± 1.4 | 62.2 ± 4.5 | 24.1 ± 3.7 |
| Det + 10AvU | 76.3 ± 6.9 | 13.7 ± 3.5 | 33.3 ± 1.7 | 58.3 ± 4.6 | 23.3 ± 4.4 |
| Det + 100AvU* | 78.6 ± 4.7 | 12.7 ± 3.0 | 34.2 ± 1.5 | 60.8 ± 4.7 | 22.4 ± 4.1 |
| Det + 1000AvU | 42.5 ± 7.2 | 12.1 ± 2.1 | 28.9 ± 1.7 | 66.1 ± 5.8 | 19.0 ± 6.2 |
| 1-6 Focal(=1)* | 77.2 ± 6.7 | 12.5 ± 2.9 | 30.6 ± 1.7 | 57.0 ± 4.6 | 20.9 ± 4.2 |
| Focal(=2) | 77.7 ± 5.2 | 12.2 ± 1.9 | 24.1 ± 0.9 | 57.5 ± 4.6 | 21.0 ± 4.1 |
| Focal(=3) | 79.0 ± 5.2 | 13.3 ± 1.6 | 18.6 ± 0.7 | 59.8 ± 4.9 | 16.6 ± 3.9 |
| 1-6 ECP(=0.1) | 78.5 ± 4.9 | 12.6 ± 2.8 | 33.5 ± 1.6 | 59.8 ± 4.9 | 22.0 ± 3.8 |
| ECP(=1.0)* | 78.8 ± 4.3 | 12.5 ± 2.6 | 36.6 ± 1.5 | 61.5 ± 4.8 | 23.2 ± 3.6 |
| ECP(=10.0) | 78.9 ± 4.5 | 12.4 ± 2.5 | 33.8 ± 1.5 | 60.1 ± 4.7 | 22.1 ± 3.5 |
| ECP(=100.0) | 62.0 ± 6.1 | 20.0 ± 1.8 | 19.9 ± 2.9 | 56.0 ± 2.8 | 36.5 ± 9.7 |
| 1-6 LS(=0.1) | 77.7 ± 6.0 | 8.9 ± 2.7 | 17.9 ± 0.3 | 57.6 ± 1.9 | 23.9 ± 4.4 |
| LS(=0.05)* | 77.7 ± 6.0 | 10.3 ± 2.9 | 24.3 ± 0.7 | 56.7 ± 3.3 | 20.6 ± 4.3 |
| LS(=0.01) | 77.9 ± 5.4 | 13.3 ± 2.8 | 31.1 ± 1.5 | 58.6 ± 3.9 | 22.4 ± 3.7 |
| 1-6 SVLS(=1)* | 78.3 ± 6.1 | 11.5 ± 3.0 | 31.4 ± 1.4 | 61.1 ± 4.9 | 23.3 ± 3.3 |
| SVLS(=2) | 79.0 ± 6.0 | 11.3 ± 2.5 | 31.4 ± 1.2 | 59.9 ± 5.4 | 21.6 ± 2.7 |
| SVLS(=3) | 78.6 ± 5.1 | 11.5 ± 2.9 | 31.1 ± 1.5 | 58.7 ± 5.0 | 22.5 ± 3.8 |
| 1-6 MbLS(,m=30) | 76.5 ± 7.1 | 13.6 ± 3.9 | 32.1 ± 2.9 | 58.9 ± 4.1 | 24.7 ± 7.7 |
| MbLS(,m=20)* | 77.5 ± 6.3 | 13.4 ± 3.0 | 33.4 ± 1.5 | 56.9 ± 5.0 | 21.5 ± 3.6 |
| MbLS(,m=10) | 76.8 ± 6.3 | 13.0 ± 3.2 | 31.7 ± 1.4 | 53.0 ± 4.5 | 20.6 ± 3.9 |
| MbLS(,m=8) | 50.3 ± 10.6 | 20.1 ± 2.8 | 26.2 ± 0.9 | 61.1 ± 7.1 | 34.1 ± 3.7 |
| [0.5pt/5pt] MbLS(,m=20) | 77.3 ± 6.2 | 13.2 ± 2.8 | 33.3 ± 1.6 | 61.0 ± 4.5 | 23.4 ± 4.1 |
| MbLS(,m=20) | 78.1 ± 5.3 | 13.0 ± 2.9 | 32.9 ± 1.5 | 57.0 ± 4.1 | 21.7 ± 3.5 |
| MbLS(,m=20) | 78.2 ± 4.9 | 12.7 ± 2.5 | 31.6 ± 1.3 | 55.0 ± 5.1 | 19.7 ± 3.5 |
| 1-6 TTA | 78.1 ± 4.6 | 12.7 ± 2.6 | 33.2 ± 1.5 | 62.7 ± 4.6 | 24.9 ± 4.1 |
| 1-6 Ens | 78.6 ± 5.2 | 10.6 ± 2.4 | 32.1 ± 1.9 | 64.7 ± 4.9 | 28.2 ± 5.1 |
| 1-6 Bayes | 75.0 ± 9.9 | 12.4 ± 4.0 | 32.2 ± 1.8 | 64.8 ± 5.0 | 27.7 ± 5.8 |
| Bayes + 10AvU | 74.9 ± 9.5 | 12.4 ± 4.0 | 32.1 ± 2.0 | 65.2 ± 4.6 | 29.1 ± 6.1 |
| Bayes + 100AvU* | 76.3 ± 7.7 | 12.1 ± 3.7 | 33.2 ± 1.7 | 65.8 ± 5.0 | 30.1 ± 6.5 |
| Bayes + 1000AvU | 75.5 ± 8.2 | 14.3 ± 4.1 | 33.5 ± 1.8 | 69.3 ± 5.6 | 32.9 ± 6.9 |
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 85.9 ± 1.8 | 14.4 ± 3.2 | 96.5 ± 0.9 | 92.6 ± 4.1 | 26.5 ± 1.5 |
| Det + 100AvU | 84.8 ± 2.3 | 16.3 ± 3.9 | 96.1 ± 0.9 | 91.7 ± 4.2 | 27.9 ± 2.8 |
| Det + 1000AvU | 84.8 ± 1.9 | 16.0 ± 3.0 | 96.4 ± 0.9 | 93.6 ± 3.2 | 29.2 ± 1.0 |
| Det + 10000AvU | 84.9 ± 3.5 | 16.7 ± 5.1 | 96.5 ± 1.0 | 91.8 ± 2.7 | 25.9 ± 2.3 |
| 1-6 Ensemble | 85.4 ± 1.7 | 13.4 ± 3.0 | 96.0 ± 1.0 | 94.8 ± 2.4 | 31.4 ± 1.6 |
| 1-6 Focal(=1) | 84.5 ± 2.7 | 13.3 ± 4.3 | 90.7 ± 1.1 | 93.0 ± 4.1 | 29.4 ± 1.7 |
| Focal(=2) | 84.4 ± 2.1 | 9.8 ± 2.6 | 82.5 ± 1.0 | 93.8 ± 2.3 | 30.9 ± 2.1 |
| Focal(=3) | 84.5 ± 1.9 | 6.4 ± 1.5 | 58.9 ± 1.3 | 92.0 ± 4.3 | 30.5 ± 2.6 |
| 1-6 ECP(=0.1) | 85.9 ± 1.8 | 14.6 ± 3.0 | 96.5 ± 0.9 | 91.9 ± 4.2 | 25.8 ± 1.7 |
| ECP(=1.0) | 85.7 ± 1.8 | 14.7 ± 3.0 | 96.4 ± 1.0 | 92.3 ± 3.9 | 26.4 ± 1.7 |
| ECP(=10.0) | 85.7 ± 1.7 | 14.8 ± 2.7 | 96.4 ± 1.0 | 91.9 ± 4.5 | 26.0 ± 1.8 |
| ECP(=100.0) | 85.7 ± 1.8 | 14.8 ± 2.8 | 96.4 ± 1.0 | 91.8 ± 4.3 | 25.8 ± 1.9 |
| ECP(=1000.0) | 86.0 ± 1.9 | 15.0 ± 3.0 | 85.0 ± 0.3 | 88.7 ± 2.1 | 26.7 ± 3.4 |
| 1-6 LS(=0.01) | 83.7 ± 2.5 | 17.2 ± 4.1 | 91.9 ± 0.9 | 85.8 ± 5.4 | 28.2 ± 2.9 |
| LS(=0.05) | 85.1 ± 1.4 | 13.6 ± 2.3 | 80.8 ± 0.9 | 84.3 ± 5.7 | 25.4 ± 2.2 |
| LS(=0.1) | 85.0 ± 2.1 | 11.1 ± 3.4 | 70.3 ± 0.6 | 85.1 ± 3.6 | 27.0 ± 2.2 |
| 1-6 SVLS(=1) | 84.5 ± 1.9 | 14.0 ± 2.6 | 92.4 ± 1.0 | 91.8 ± 2.3 | 22.9 ± 1.8 |
| SVLS(=2) | 85.0 ± 1.8 | 12.9 ± 3.1 | 92.4 ± 0.9 | 91.4 ± 3.0 | 22.1 ± 1.4 |
| SVLS(=3) | 85.0 ± 1.6 | 13.1 ± 2.7 | 92.1 ± 0.9 | 91.2 ± 2.5 | 21.9 ± 1.4 |
| 1-6 MbLS(,m=10) | 84.8 ± 1.4 | 17.5 ± 5.1 | 95.7 ± 1.1 | 91.2 ± 4.1 | 31.1 ± 1.7 |
| MbLS(,m=8) | 83.8 ± 1.3 | 16.0 ± 2.2 | 93.9 ± 0.9 | 90.5 ± 3.5 | 27.9 ± 2.1 |
| MbLS(,m=5) | 84.3 ± 1.6 | 15.5 ± 2.8 | 90.4 ± 0.8 | 90.1 ± 5.6 | 28.2 ± 2.2 |
| MbLS(,m=3) | 84.2 ± 2.1 | 12.8 ± 3.3 | 70.8 ± 0.4 | 82.1 ± 3.5 | 28.8 ± 5.6 |
| [0.5pt/5pt] MbLS(,m=10) | 83.7 ± 1.2 | 17.4 ± 4.4 | 96.0 ± 1.0 | 91.2 ± 3.9 | 30.5 ± 1.5 |
| MbLS(,m=10) | 83.9 ± 1.5 | 17.8 ± 4.1 | 95.0 ± 1.2 | 90.8 ± 3.6 | 30.9 ± 1.6 |
| 1-6 TTA | 85.6 ± 1.7 | 14.5 ± 3.1 | 96.3 ± 0.9 | 92.5 ± 4.0 | 27.2 ± 1.6 |
| 1-6 Bayes | 85.7 ± 2.3 | 10.7 ± 3.0 | 93.2 ± 1.8 | 95.3 ± 1.9 | 30.3 ± 2.9 |
| Bayes + 100AvU | 86.1 ± 3.0 | 11.5 ± 3.8 | 94.9 ± 2.1 | 95.9 ± 2.0 | 31.5 ± 3.5 |
| Bayes + 1000AvU | 85.8 ± 2.8 | 12.0 ± 3.9 | 95.5 ± 1.9 | 96.3 ± 2.4 | 32.0 ± 3.3 |
| Bayes + 10000AvU | 86.0 ± 2.4 | 10.9 ± 3.0 | 96.1 ± 1.7 | 93.1 ± 2.1 | 29.3 ± 3.1 |
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 84.1 ± 5.6 | 12.9 ± 6.0 | 96.1 ± 3.4 | 92.5 ± 5.7 | 28.0 ± 3.7 |
| Det + 100AvU | 83.7 ± 6.7 | 16.6 ± 7.2 | 95.7 ± 3.3 | 91.6 ± 6.2 | 27.2 ± 2.9 |
| Det + 1000AvU* | 83.7 ± 6.8 | 16.9 ± 8.1 | 95.9 ± 3.8 | 92.1 ± 6.8 | 28.2 ± 3.4 |
| Det + 10000AvU | 83.4 ± 6.4 | 18.1 ± 7.9 | 96.1 ± 3.7 | 90.7 ± 5.6 | 26.1 ± 3.4 |
| 1-6 Focal (=1)* | 81.1 ± 15.4 | 10.2 ± 5.0 | 90.3 ± 0.3 | 93.2 ± 5.5 | 29.3 ± 3.4 |
| Focal (=2) | 83.1 ± 6.2 | 10.4 ± 6.8 | 81.6 ± 2.5 | 92.9 ± 5.3 | 30.1 ± 3.7 |
| Focal (=3) | 82.3 ± 7.2 | 8.0 ± 6.4 | 58.7 ± 1.2 | 92.5 ± 5.4 | 31.8 ± 3.5 |
| 1-6 ECP (=0.1) | 84.1 ± 5.4 | 16.5 ± 7.0 | 96.1 ± 3.4 | 92.3 ± 6.0 | 27.6 ± 3.9 |
| ECP (=1.0) | 84.1 ± 5.5 | 16.4 ± 7.0 | 96.1 ± 3.3 | 92.3 ± 6.0 | 27.8 ± 4.3 |
| ECP (=10.0)* | 84.0 ± 5.5 | 16.7 ± 7.1 | 96.1 ± 3.4 | 92.1 ± 6.0 | 27.6 ± 4.3 |
| ECP (=100.0) | 84.0 ± 5.5 | 16.6 ± 7.0 | 96.0 ± 3.0 | 92.1 ± 6.0 | 27.6 ± 4.1 |
| ECP (=1000.0) | 84.1 ± 5.7 | 16.6 ± 7.0 | 86.1 ± 3.2 | 92.2 ± 5.9 | 27.5 ± 4.3 |
| 1-6 LS (=0.01) | 82.5 ± 8.3 | 18.0 ± 9.4 | 91.3 ± 3.9 | 86.2 ± 7.9 | 27.0 ± 3.8 |
| LS (=0.05)* | 83.4 ± 7.2 | 15.1 ± 8.6 | 80.4 ± 2.9 | 83.2 ± 7.8 | 25.1 ± 3.1 |
| LS (=0.1) | 84.1 ± 5.6 | 11.6 ± 7.0 | 70.1 ± 1.8 | 84.7 ± 6.2 | 26.9 ± 3.3 |
| 1-6 SVLS (=1) | 83.4 ± 7.1 | 14.7 ± 8.8 | 92.0 ± 3.7 | 90.9 ± 7.4 | 22.9 ± 2.9 |
| SVLS (=2)* | 83.5 ± 6.7 | 14.0 ± 8.1 | 91.9 ± 4.1 | 90.5 ± 7.9 | 21.7 ± 2.6 |
| SVLS(=3) | 83.2 ± 8.1 | 14.3 ± 9.7 | 91.5 ± 3.9 | 91.0 ± 6.8 | 23.1 ± 3.1 |
| 1-6 MbLS (,m=3) | 83.2 ± 6.3 | 13.3 ± 7.8 | 70.6 ± 1.7 | 82.2 ± 6.3 | 27.7 ± 3.4 |
| MbLS (,m=5) | 82.8 ± 6.6 | 16.7 ± 8.0 | 89.9 ± 3.2 | 90.5 ± 7.2 | 27.2 ± 4.4 |
| MbLS (,m=8) | 83.5 ± 5.8 | 17.1 ± 7.0 | 95.3 ± 3.6 | 93.0 ± 5.2 | 27.8 ± 4.1 |
| MbLS (,m=10) | 84.2 ± 5.3 | 18.1 ± 6.1 | 95.5 ± 3.3 | 91.7 ± 6.1 | 26.5 ± 3.5 |
| [0.5pt/5pt] MbLS(,m=10)* | 84.2 ± 4.9 | 17.9 ± 7.4 | 95.6 ± 2.9 | 92.2 ± 5.6 | 26.9 ± 3.6 |
| MbLS(,m=10) | 83.9 ± 5.2 | 17.9 ± 8.0 | 95.1 ± 3.2 | 91.9 ± 5.9 | 26.2 ± 4.1 |
| 1-6 TTA | 83.8 ± 5.8 | 16.4 ± 7.1 | 96.0 ± 3.5 | 92.7 ± 5.6 | 28.8 ± 3.9 |
| 1-6 Ensemble | 84.5 ± 5.7 | 11.3 ± 6.5 | 95.2 ± 3.5 | 94.3 ± 4.3 | 30.0 ± 4.6 |
| 1-6 Bayes | 84.0 ± 5.8 | 8.6 ± 4.7 | 92.1 ± 2.6 | 94.7 ± 3.1 | 29.1 ± 4.8 |
| Bayes + 100AvU | 84.1 ± 6.4 | 12.0 ± 6.2 | 94.4 ± 3.1 | 95.5 ± 2.9 | 28.9 ± 5.0 |
| Bayes + 1000AvU* | 84.9 ± 6.9 | 8.9 ± 6.0 | 94.5 ± 3.2 | 95.7 ± 3.2 | 30.5 ± 4.5 |
| Bayes + 10000AvU | 85.2 ± 5.9 | 11.0 ± 6.3 | 94.2 ± 3.6 | 95.9 ± 3.5 | 30.2 ± 4.0 |
| Experiment | DICE (x) | ECE (x) | AvU-AUC (x) | ROC-AUC (x) | PRC-AUC (x) |
|---|---|---|---|---|---|
| Det | 74.2 ± 12.6 | 15.6 ± 6.3 | 92.3 ± 5.4 | 87.9 ± 7.5 | 22.1 ± 6.2 |
| Det + 100AvU | 74.2 ± 13.3 | 23.6 ± 11.2 | 93.0 ± 4.2 | 87.1 ± 6.2 | 22.2 ± 5.7 |
| Det + 1000AvU* | 74.5 ± 13.0 | 27.6 ± 14.3 | 92.2 ± 5.7 | 88.2 ± 7.6 | 22.0 ± 7.1 |
| Det + 10000AvU | 72.7 ± 15.1 | 27.6 ± 14.3 | 92.4 ± 5.2 | 82.3 ± 9.4 | 19.6 ± 6.2 |
| 1-6 Focal(=1)* | 71.2 ± 17.4 | 12.1 ± 5.8 | 85.4 ± 6.1 | 89.0 ± 7.1 | 24.3 ± 6.7 |
| Focal(=2) | 76.7 ± 10.8 | 12.8 ± 8.2 | 72.0 ± 9.3 | 87.2 ± 7.6 | 22.4 ± 6.4 |
| Focal(=3) | 73.2 ± 13.7 | 11.6 ± 7.7 | 49.7 ± 9.4 | 87.1 ± 8.5 | 27.0 ± 7.2 |
| 1-6 ECP(=0.1) | 74.6 ± 12.5 | 22.8 ± 10.5 | 92.1 ± 5.5 | 87.6 ± 7.6 | 21.3 ± 6.6 |
| ECP(=1.0) | 73.9 ± 13.1 | 23.2 ± 10.7 | 91.9 ± 5.6 | 87.2 ± 7.2 | 21.2 ± 6.4 |
| ECP(=10.0)* | 74.8 ± 12.5 | 22.3 ± 10.2 | 91.6 ± 6.3 | 87.2 ± 8.1 | 20.6 ± 7.0 |
| ECP(=100.0) | 74.9 ± 12.3 | 22.7 ± 10.5 | 92.1 ± 5.5 | 87.7 ± 8.0 | 21.5 ± 7.2 |
| ECP(=1000.0) | 74.6 ± 12.5 | 22.7 ± 10.3 | 92.2 ± 5.6 | 87.6 ± 7.7 | 21.5 ± 6.7 |
| 1-6 LS(=0.01) | 71.6 ± 15.1 | 24.6 ± 11.6 | 87.9 ± 5.3 | 84.3 ± 7.5 | 22.7 ± 6.2 |
| LS(=0.05)* | 74.5 ± 13.0 | 21.7 ± 11.5 | 77.2 ± 4.6 | 79.5 ± 8.9 | 19.1 ± 7.2 |
| LS(=0.1) | 75.2 ± 12.2 | 18.1 ± 10.1 | 67.4 ± 3.8 | 79.0 ± 8.4 | 19.9 ± 6.4 |
| 1-6 SVLS(=1) | 74.9 ± 11.7 | 19.7 ± 9.1 | 88.5 ± 5.5 | 87.2 ± 7.4 | 18.7 ± 5.1 |
| SVLS(=2)* | 76.9 ± 11.5 | 17.9 ± 9.3 | 88.3 ± 5.2 | 87.2 ± 7.2 | 16.4 ± 5.2 |
| SVLS(=3) | 74.3 ± 13.5 | 21.4 ± 12.6 | 88.4 ± 5.1 | 86.3 ± 8.2 | 19.4 ± 5.0 |
| 1-6 MbLS(, m=10) | 72.3 ± 15.9 | 20.9 ± 7.9 | 91.4 ± 5.7 | 87.9 ± 6.9 | 22.2 ± 6.7 |
| MbLS(, m=8) | 74.1 ± 13.5 | 20.7 ± 8.7 | 88.3 ± 8.2 | 85.0 ± 10.4 | 18.8 ± 8.8 |
| MbLS(, m=5) | 74.7 ± 13.3 | 22.0 ± 11.3 | 86.9 ± 5.0 | 87.1 ± 7.9 | 22.0 ± 6.4 |
| MbLS(, m=3) | 74.0 ± 13.3 | 20.5 ± 11.7 | 68.6 ± 2.9 | 78.0 ± 7.2 | 21.5 ± 6.7 |
| [0.5pt/5pt] MbLS(, m=10)* | 73.6 ± 12.5 | 19.9 ± 7.4 | 91.8 ± 3.4 | 86.5 ± 7.2 | 21.8 ± 5.6 |
| MbLS(, m=10) | 72.1 ± 16.1 | 20.2 ± 6.7 | 91.4 ± 5.5 | 86.5 ± 9.0 | 22.2 ± 6.7 |
| 1-6 TTA | 74.0 ± 12.8 | 23.7 ± 11.4 | 92.8 ± 4.8 | 88.6 ± 7.4 | 24.9 ± 5.8 |
| 1-6 Ensemble | 76.3 ± 12.2 | 9.7 ± 5.0 | 89.9 ± 6.6 | 91.6 ± 5.2 | 28.4 ± 5.7 |
| 1-6 Bayes | 70.6 ± 16.6 | 11.8 ± 7.2 | 86.2 ± 6.0 | 89.1 ± 7.4 | 25.7 ± 5.1 |
| Bayes + 100AvU | 72.1 ± 14.4 | 20.0 ± 11.8 | 91.0 ± 3.8 | 92.7 ± 4.0 | 30.2 ± 6.5 |
| Bayes + 1000AvU* | 76.3 ± 12.6 | 11.4 ± 6.7 | 89.5 ± 6.2 | 90.6 ± 6.9 | 26.2 ± 7.4 |
| Bayes + 10000AvU | 76.6 ± 12.7 | 17.1 ± 10.1 | 88.6 ± 6.5 | 90.4 ± 6.3 | 23.3 ± 7.4 |
Appendix D Visual Results
Visual results in Figure 2 and Figure 3 show pairs of consecutive CT/MR slices to better understand the 3D nature of the output uncertainty across all models. We show examples with both high and low DICE to investigate the presence and absence of uncertainty in different regions of the model prediction.
D.1 Head-And-Neck CT
The first two rows of Figure 2a and Figure 2b show the mandible (i.e. lower jaw bone) with only the Bayes+AvU model having overall low uncertainty in accurate regions and high uncertainty in (or close to) inaccurate regions.
In the next set of rows for head-and-necks CTs, we observe the parotid gland, a salivary organ, with (Figure 2a - Case 2) and without (Figure 2b - Case 2, Case 3) a dental scattering issue. In both cases, while the Det model shows low uncertainty, the baseline Bayes model shows high uncertainty in accurate regions. Usage of the AvU loss lowers uncertainty in these regions, while still exhibiting uncertainty in the erroneous regions, for e.g. the medial (i.e. internal) portion of the organ in Figure 2a (Case 2).
Moving on to our last case, we see the submandibular gland, another salivary gland in Figure 2a (Case 3). The Ensemble, Focal, SVLS and MBLS models all display high uncertainty in the core of the organ, which are also accurately predicted. On the other hand, the AvU loss minimizes the uncertainty and shows uncertainty in the erroneous region on the second slice.
D.2 Prostate MR
For the prostate datasets, we see two cases with high DICE in Figure 3a (Case 1) and Figure 3b (Case 2) where the use of the AvU loss reduces uncertainty for the baseline Bayes model.
We also see cases with low DICE in Figure 3a (Case 2) and Figure 3b (Case 1). Due to their low DICE all models display high uncertainty, but the Bayes+AvU model shows high overlap between its uncertain and erroneous regions. The same is also observed in Figure 3b (Case 3).
Finally, in Figure 3a (Case 3), we do not see any clear benefit of using the AvU loss on the Bayes model.
Appendix E BayesH model
| TestDataset | Model | DICE (x) | ECE (x) | ROC-AUC (x) | PRC-AUC(x) |
|---|---|---|---|---|---|
| ID————H&N CT(RTOG) | Det | 84.2 ± 2.7 | 9.0 ± 2.1 | 73.0 ± 5.7 | 21.0 ± 4.8 |
| Ensemble | 85.0 ± 2.6 | 8.6 ± 2.1 | 78.6 ± 4.7 | 25.7 ± 6.8 | |
| 2-6 | Bayes | 83.9 ± 2.6 | 8.6 ± 2.1 | 74.1 ± 5.4 | 22.1 ± 3.5 |
| Bayes+AvU | 83.6 ± 2.5 | 7.6 ± 2.5† | 76.1 ± 5.6† | 25.1 ± 5.3† | |
| 2-6 | BayesH | 83.6 ± 2.9 | 9.2 ± 2.6 | 70.4 ± 7.0 | 20.1 ± 3.8 |
| BayesH+AvU | 84.1 ± 2.7 | 8.4 ± 2.4† | 74.1 ± 5.4† | 21.3 ± 4.6† | |
| OOD————H&N CT(STRSeg) | Det | 78.1 ± 4.6 | 12.9 ± 2.6 | 62.2 ± 4.5 | 24.1 ± 3.7 |
| Ensemble | 78.6 ± 5.2 | 10.6 ± 2.4 | 64.7 ± 4.9 | 28.2 ± 5.1 | |
| 2-6 | Bayes | 75.0 ± 9.9 | 12.4 ± 4.0 | 64.8 ± 5.0 | 27.7 ± 5.8 |
| Bayes+AvU | 76.3 ± 7.6† | 12.1 ± 3.7 | 65.8 ± 5.0† | 30.1 ± 6.5† | |
| 2-6 | BayesH | 77.5 ± 6.6 | 12.6 ± 3.3 | 61.1 ± 4.1 | 23.5 ± 4.7 |
| BayesH+AvU | 78.8 ± 5.1† | 12.1 ± 3.2 | 64.8 ± 3.8† | 23.8 ± 4.0† | |
| ID————ProstateMR(PrMedDec) | Det | 84.1 ± 5.6 | 12.9 ± 6.0 | 92.5 ± 5.7 | 28.0 ± 3.7 |
| Ensemble | 84.5 ± 5.7 | 11.3 ± 6.5 | 94.3 ± 4.3 | 30.0 ± 4.6 | |
| 2-6 | Bayes | 84.0 ± 5.8 | 8.6 ± 4.7 | 94.7 ± 3.1 | 29.1 ± 4.8 |
| Bayes+AvU | 84.9 ± 6.9 | 8.9 ± 6.0 | 95.7 ± 3.2† | 30.5 ± 4.5† | |
| 2-6 | BayesH | 82.3 ± 5.2 | 9.3 ± 4.3 | 93.6 ± 2.9 | 28.4 ± 4.2 |
| BayesH+AvU | 84.5 ± 6.3† | 9.4 ± 6.5 | 94.9 ± 3.1† | 30.1 ± 4.9† | |
| OOD————ProstateMR(PR12) | Det | 74.2 ± 12.6 | 15.6 ± 6.3 | 87.9 ± 7.5 | 22.1 ± 6.2 |
| Ensemble | 76.3 ± 12.2 | 9.7 ± 5.0 | 91.6 ± 5.2 | 28.4 ± 5.7 | |
| 2-6 | Bayes | 70.6 ± 16.6 | 11.8 ± 7.2 | 89.1 ± 7.4 | 25.7 ± 5.1 |
| Bayes+AvU | 76.3 ± 12.6† | 11.4 ± 6.7† | 90.6 ± 6.9† | 26.2 ± 7.4† | |
| 2-6 | BayesH | 71.3 ± 14.4 | 12.1 ± 6.7 | 88.9 ± 6.3 | 25.1 ± 4.9 |
| BayesH+AvU | 74.1 ± 13.8† | 11.9 ± 6.2† | 89.9 ± 6.2† | 25.9 ± 5.4† |