ContourDiff: Unpaired Medical Image Translation with Structural Consistency
Yuwen Chen1 , Nicholas Konz1, Hanxue Gu1, Haoyu Dong1, Yaqian Chen1, Lin Li2, Jisoo Lee3, Maciej A. Mazurowski1,2,3,4
, Nicholas Konz1, Hanxue Gu1, Haoyu Dong1, Yaqian Chen1, Lin Li2, Jisoo Lee3, Maciej A. Mazurowski1,2,3,4
1: Department of Electrical and Computer Engineering, Duke University, Durham, NC 27708, 2: Department of Biostatistics & Bioinformatics, Duke University, Durham, NC 27708, 3: Department of Radiology, Duke University, Durham, NC 27708, 4: Department of Computer Science, Duke University, Durham, NC, 27708
Publication date: 2025/11/25
https://doi.org/10.59275/j.melba.2025-79a2
Abstract
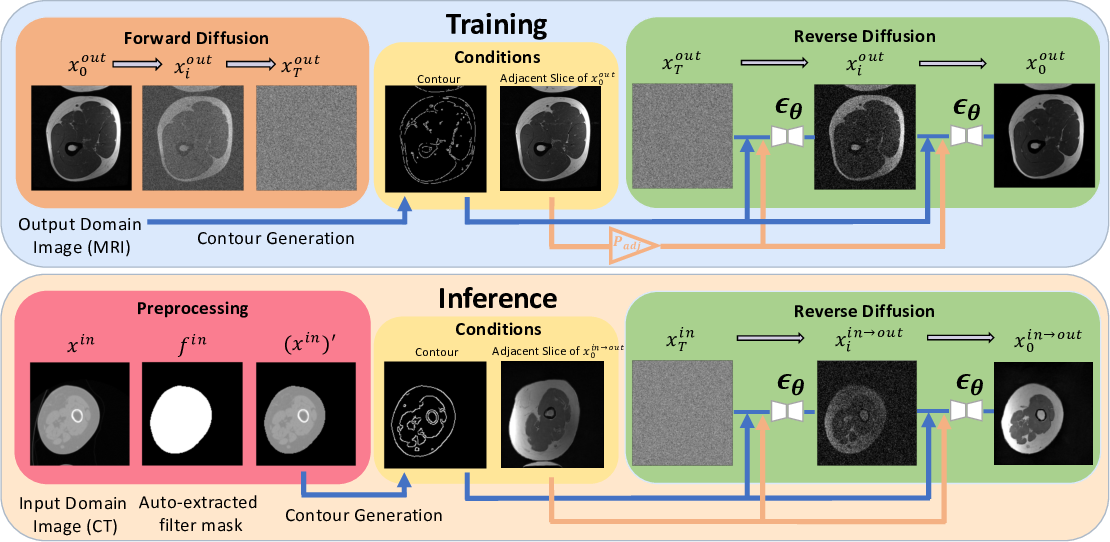
Accurately translating medical images between different modalities, such as Computed Tomography (CT) to Magnetic Resonance Imaging (MRI), has numerous downstream clinical and machine learning applications. While several methods have been proposed to achieve this, they often prioritize perceptual quality with respect to output domain features over preserving anatomical fidelity. However, maintaining anatomy during translation is essential for many tasks, e.g., when leveraging masks from the input domain to develop a segmentation model with images translated to the output domain. To address these challenges, we propose ContourDiff with Spatially Coherent Guided Diffusion (SCGD), a novel framework that leverages domain-invariant anatomical contour representations of images. These representations are simple to extract from images, yet form precise spatial constraints on their anatomical content. We introduce a diffusion model that converts contour representations of images from arbitrary input domains into images in the output domain of interest. By applying the contour as a constraint at every diffusion sampling step, we ensure the preservation of anatomical content. We evaluate our method on challenging lumbar spine and hip-and-thigh CT-to-MRI translation tasks, via (1) the performance of segmentation models trained on translated images applied to real MRIs, and (2) the foreground FID and KID of translated images with respect to real MRIs. Our method outperforms other unpaired image translation methods by a significant margin across almost all metrics and scenarios. Moreover, it achieves this without the need to access any input domain information during training and we further verify its zero-shot capability, showing that a model trained on one anatomical region can be directly applied to unseen regions without retraining. Our code is available at https://github.com/mazurowski-lab/ContourDiff
Keywords
Unpaired Image Translation · Medical Image Segmentation · Diffusion Model
Bibtex
@article{melba:2025:031:chen,
title = "ContourDiff: Unpaired Medical Image Translation with Structural Consistency",
author = "Chen, Yuwen and Konz, Nicholas and Gu, Hanxue and Dong, Haoyu and Chen, Yaqian and Li, Lin and Lee, Jisoo and Mazurowski, Maciej A.",
journal = "Machine Learning for Biomedical Imaging",
volume = "3",
issue = "November 2025 issue",
year = "2025",
pages = "711--727",
issn = "2766-905X",
doi = "https://doi.org/10.59275/j.melba.2025-79a2",
url = "https://melba-journal.org/2025:031"
}
RIS
TY - JOUR
AU - Chen, Yuwen
AU - Konz, Nicholas
AU - Gu, Hanxue
AU - Dong, Haoyu
AU - Chen, Yaqian
AU - Li, Lin
AU - Lee, Jisoo
AU - Mazurowski, Maciej A.
PY - 2025
TI - ContourDiff: Unpaired Medical Image Translation with Structural Consistency
T2 - Machine Learning for Biomedical Imaging
VL - 3
IS - November 2025 issue
SP - 711
EP - 727
SN - 2766-905X
DO - https://doi.org/10.59275/j.melba.2025-79a2
UR - https://melba-journal.org/2025:031
ER -