1 Introduction
In the times of the rapidly emerging coronavirus disease 2019 (COVID-19), hot spots and growing concerns about a second wave are making it crucial to streamline patient diagnosis and management. Many experts in the medical community believe that artificial intelligence (AI) systems could lessen the burden on hospitals dealing with outbreaks by processing imaging data [Kim, 2020; Rubin et al., 2020]. Hospitals have deployed AI-driven computed tomography (CT) scan interpreters in China [Simonite, 2020] and Italy [Lyman, 2020], as well as AI initiatives to improve triaging of COVID-19 patients (i.e., discharge, general admission, or ICU care) and allocation of hospital resources [Strickland, 2020; Hao, 2020].
Carefully curated and annotated data is the first step to developing any diagnostic or management tool. While there exist large public datasets of more typical chest X-rays (CXR) [Wang et al., 2017; Bustos et al., 2019; Irvin et al., 2019; Johnson et al., 2019; Demner-Fushman et al., 2016], there was no public collection of COVID-19 CXR or CT scans designed to be used for computational analysis. We first made data public in mid February 2020 and the dataset has rapidly grown [Cohen et al., 2020c]. More recently, in June, the BIMCV COVID-19+ dataset [De La Iglesia Vayá et al., 2020] was released. While it has more samples than the dataset we present, we complement their work with a focus on prospective metadata from multiple medical centers and countries.
Many physicians remain reluctant to share their patients’ anonymized imaging data in open datasets, even after obtaining consent, due to ethical concerns over privacy and confidentiality and a hospital culture that, in our experience, does not reward sharing [Keen et al., 2013; Lee and Yoon, 2017; Kostkova et al., 2016; Floca, 2014]. In order to access data in one hospital, researchers must submit a protocol to the hospital’s institutional review board (IRB) for approval and build their own data management system. While this is important for patient safety, such routines must be repeated for every hospital, resulting in a lengthy bureaucratic process that hurts reproducibility and external validation.
The ultimate goal of this project is to aggregate all publicly available radiographs, including papers and other datasets that can be combined. For research articles, images are extracted by hand, while for websites, such as Radiopaedia and Eurorad, data collection is partially automated using scrapers that extract a subset of the metadata while we hand review case notes to determine clinical events. This work provides three primary contributions:
- •
We create the first public COVID-19 CXR image data collection, currently totalling 679 frontal chest X-ray images from 412 people from 26 countries and growing. The dataset contains clinical attributes about survival, ICU stay, intubation events, blood tests, location, and freeform clinical notes for each image/case. In contrast to other works, we focus on prospective metadata for the development of prognostic and management tools from CXR. Images collected have already been made public and are presented in an ML-ready dataset with toolchains that are easily used in many testable settings.
- •
We also present and discuss clinical use-cases and propose ML tasks that may address these clinical use cases such as predicting pneumonia severity, survival outcome, and need for the intensive care unit (ICU). Benchmark results using transfer learning with neural networks are included.
- •
We also discuss how to use the location information in this dataset for a Leave-One-Country/Continent-Out (LOCO) evaluation to simulate domain shift and provide a more robust evaluation.
Currently, all images and data are released under the following GitHub URL111https://github.com/ieee8023/covid-chestxray-dataset. We hope that this dataset and tasks will allow for quick uptake of COVID-related prediction problems in the machine learning community.
2 Background and Related Work
2.1 Learning in Medical Imaging
In recent years, ML applications on CXR data have seen rising interest, such as lung segmentation [Gordienko et al., 2018; Islam and Zhang, 2018], tuberculosis and cancer analysis [Gordienko et al., 2018; Stirenko et al., 2018; Lakhani and Sundaram, 2017], abnormality detection [Islam et al., 2017], explanation [Singla et al., 2020], and multi-modality predictions Rubin et al. [2018]; Hashir et al. [2020]. With the availability of large-scale public CXR datasets created with ML in mind (e.g. CheXpert [Irvin et al., 2019], Chest-xray8 [Wang et al., 2017], PadChest [Bustos et al., 2019] or MIMIC-CXR [Johnson et al., 2019]), neural networks have even reported being able to achieve performance near radiologist levels [Rajpurkar et al., 2018, 2017; Irvin et al., 2019; Putha et al., 2018a; Majkowska et al., 2019; Putha et al., 2018b]. However, the adoption has its own challenges [Couzin-Frankel, 2019].
2.2 Use of Imaging for COVID-19
In this section we will give a high level overview of the recommendations for the clinical use of imaging in COVID-19 patient management. It should be noted that this list is only meant to help guide algorithm development and is not medically comprehensive.
Ever since the dawn of the outbreak, imaging has stood out as a promising avenue of research and care [Zu et al., 2020; Poggiali et al., 2020]. Particularly in the beginning of the outbreak, computed tomography (CT) scans captured the attention of both the medical [Ng et al., 2020; Kanne et al., 2020] and the ML [McCall, 2020] communities.
From March to May 2020, the Fleischner Society [Rubin et al., 2020], American College of Radiology (ACR) [ACR, 2020], Canadian Association of Radiologists (CAR) [Dennie et al., 2020a], Canadian Society of Thoracic Radiology (CSTR) Dennie et al. [2020b], and British Society of Thoracic Imaging (BSTI) [Nair et al., 2020] released the following recommendations:
- 1.
Imaging tests (CXR and chest CT scans) should not be used alone to diagnose COVID-19 nor used systematically on all patients with suspected COVID-19;
- 2.
Findings on CT scans and CXR are non-specific and these imaging techniques should not be used to inform decisions on whether to test a patient for COVID-19 (in other words, normal chest imaging results do not exclude the possibility of COVID-19 infection and abnormal chest imaging findings are not specific for diagnosis);
- 3.
CXR and chest CT scans can be used for patients at risk of disease progression and with worsening respiratory status as well as in resource-constrained environments for triage of patients with suspected COVID-19.
However, CXRs remain the first choice in terms of the initial imaging test when caring for patients with suspected COVID-19. CXRs are the preferred initial imaging modality when pneumonia is suspected [ACR, 2018] and the radiation dose of CXR (0.02 mSv for a PA film) is lower than the radiation dose of chest CT scans (7 mSv), putting the patients less at risk of radiation-related diseases such as cancer [FDA, 2017].
In addition, CXR are cheaper than CT scans, making them more viable financially for healthcare systems and patients. [Beek, 2015; Ball et al., 2017]. Finally, portable CXR units can be wheeled into ICU as well as emergency rooms (ER) and are easily cleaned afterwards, reducing impact on patient flow and risks of infection [Dennie et al., 2020a; ACR, 2020].
A recent hypothesis that is increasingly confirmed by research suggesting that COVID-19 is not a pulmonary but rather a vascular disease [Varga et al., 2020] [Ackermann et al., 2020]. This could explain the variety of symptoms which do not necessarily include pneumonia but can still lead to ICU hospitalization [Wadman et al., 2020].
2.3 COVID-19 Prediction from CXR
While many works have attempted to predict COVID-19 through medical imaging, the results have been on small or private data. Many of these methods use the dataset presented here. For example, while promising results were achieved by Raj [2020] (90% AUC), these were reported over a large private dataset and are not reproducible. Additionally, much research is presented without appropriate evaluations, potentially leading to overfitting and performance overestimation [Maguolo and Nanni, 2020; Tartaglione et al., 2020; DeGrave et al., 2020]. Many prediction models are not viable for use in clinical practice, as they are inadequately reported, particularly with regard to their performance, and at strong risk of bias Wynants et al. [2020]. A study by Murphy et al. [2020] found that an AI system achieved an AUC of 0.81 and was comparable with that of six independent readers.
Most ML work using this dataset has focused on COVID-19 Prediction similar to our task definition in §5.1. Many have used a transfer learning approach, similar to what we use for baselines in this work, where a model that is pretrained on existing CXR datasets is used to construct features from the images in this dataset which are lower dimensional and are less prone to overfitting [Apostolopoulos and Mpesiana, 2020][Minaee et al., 2020]. Another similar approach taken is utilizing a multi-task network which predicts both standard CXR tasks as well as a COVID-19 task to assign a likelihood to samples and perform anomaly detection [Zhang et al., 2020]. Another work has utilized the pathology hierarchy we provide to improve predictions by using the Clus-HMC method [Pereira et al., 2020].
Many groups have also used this dataset to make severity and survival outcome predictions similar to our task definition in §5.2 and 5.3. Such work like Signoroni et al. [2020] focused on predicting a Brixia Score which has been clinically studied against severity and outcomes [Borghesi and Maroldi, 2020b]. It is laborious to create these annotations so an approach by Amer et al. [2020] predicts a lung segmentation, generates a prediction saliency, and then calculates the ratio of coverage which acts as a proxy for the Brixia and geographic extent scores we discuss later.
| Type | Genus or Species | Image Count |
|---|---|---|
| Viral | COVID-19 (SARSr-CoV-2) | 468 |
| SARS (SARSr-CoV-1) | 16 | |
| MERS-CoV | 10 | |
| Varicella | 5 | |
| Influenza | 4 | |
| Herpes | 3 | |
| Bacterial | Streptococcus spp. | 13 |
| Klebsiella spp. | 9 | |
| Escherichia coli | 4 | |
| Nocardia spp. | 4 | |
| Mycoplasma spp. | 5 | |
| Legionella spp. | 7 | |
| Unknown | 2 | |
| Chlamydophila spp. | 1 | |
| Staphylococcus spp. | 1 | |
| Fungal | Pneumocystis spp. | 24 |
| Aspergillosis spp. | 2 | |
| Lipoid | Not applicable | 8 |
| Aspiration | Not applicable | 1 |
| Unknown | Unknown | 59 |
3 Cohort Details
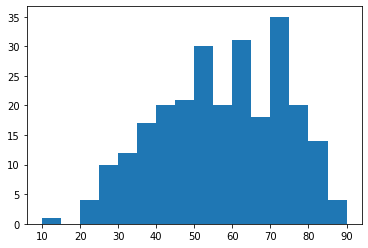
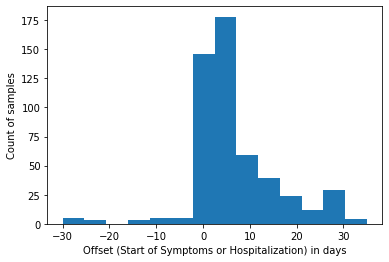
The current statistics as of September 18th 2020 are shown in Table 3, which presents the distribution of frontal CXR by diagnosis, types of pneumonia and responsible microorganisms when applicable. For each image, attributes shown in Table 1 are collected. Statistics are presented by sub-region in Table 2 and by projection/view in Table 3. Figures 3 and 3 present demographics for patients and frontal CXR images. In terms of unique patients the M/F ratio is 230/139. In total, 761 images were collected, of which 679 are frontal and 82 are lateral view. Of the frontal views 518 are standard frontal PA/AP (Posteroanterior/Anteroposterior) views and 161 are AP Supine (Anteroposterior laying down). The images originate from various hospitals across 26 different countries.
| Attribute | Description |
|---|---|
| patientid | Internal identifier |
| offset | Number of days since the start of symptoms or hospitalization for each image. If a report indicates "after a few days", then 5 days is assumed. |
| sex | Male (M), Female (F), or blank |
| age | Age of the patient in years |
| finding | Type of pneumonia |
| RT_PCR_positive | Yes (Y) or no (N) or blank if not reported/taken |
| survival | If the patient survived the disease. Yes (Y) or no (N) |
| intubated | Yes (Y) if the patient was intubated (or ventilated) at any point during this illness or No (N) or blank if unknown. |
| went_icu | Yes (Y) if the patient was in the ICU (intensive care unit) or CCU (critical care unit) at any point during this illness or No (N) or blank if unknown. |
| needed_supplemental_O2 | Yes (Y) if the patient required supplemental oxygen at any point during this illness or No (N) or blank if unknown. |
| extubated | Yes (Y) if the patient was successfully extubated or No (N) or blank if unknown. |
| temperature | Temperature of the patient in Celsius at the time of the image. |
| view | Posteroanterior (PA), Anteroposterior (AP), AP Supine (APS), or Lateral (L) for X-rays; Axial or Coronal for CT scans |
| modality | CT, X-ray, or something else |
| date | Date on which the image was acquired |
| location | Hospital name, city, state, country |
| filename | Name with extension |
| doi | Digital object identifier (DOI) of the research article |
| url | URL of the paper or website where the image came from |
| license | License of the image such as CC BY-NC-SA. Blank if unknown |
| clinical_notes | Clinical notes about the image and/or the patient |
| other_notes | e.g. credit |
| Frontal | Went | RT-PCR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | CXR | Age | Survival | ICU | Intubation | COVID-19 | Positive | Male | ||
| Region | Sub-region | Count | Count | Mean | Y/N | Y/N | Y/N | Count | Count | Ratio |
| Africa | Northern Africa | 5 | 13 | 40.00 | 0/1 | 0/0 | 0/0 | 0 | 0 | 0.00 |
| Americas | Latin Amer./Caribbean | 5 | 5 | 42.80 | 1/0 | 1/1 | 0/1 | 3 | 1 | 0.80 |
| Northern America | 27 | 46 | 53.12 | 6/5 | 11/2 | 11/4 | 17 | 15 | 0.56 | |
| Asia | Eastern Asia | 54 | 90 | 55.14 | 16/6 | 6/5 | 8/6 | 50 | 40 | 0.35 |
| South-eastern Asia | 9 | 17 | 46.67 | 4/1 | 1/3 | 1/3 | 8 | 8 | 0.56 | |
| Southern Asia | 15 | 24 | 48.27 | 4/0 | 3/0 | 3/0 | 14 | 14 | 0.73 | |
| Western Asia | 13 | 20 | 46.92 | 2/0 | 3/0 | 2/0 | 6 | 4 | 0.54 | |
| Europe | Eastern Europe | 4 | 7 | 67.50 | 1/0 | 0/0 | 0/0 | 1 | 1 | 0.50 |
| Northern Europe | 31 | 60 | 58.78 | 3/3 | 7/0 | 7/1 | 22 | 18 | 0.55 | |
| Southern Europe | 123 | 195 | 59.30 | 31/4 | 25/9 | 13/22 | 91 | 74 | 0.63 | |
| Western Europe | 53 | 92 | 52.83 | 19/2 | 19/29 | 1/1 | 49 | 5 | 0.60 | |
| Oceania | Australia/New Zealand | 42 | 60 | 50.48 | 4/0 | 1/1 | 1/1 | 1 | 2 | 0.57 |
| Unknown | Unknown | 31 | 50 | 53.64 | 3/3 | 6/0 | 7/2 | 19 | 0 | 0.55 |
| Totals | 412 | 679 | 55.1 | 119 | 133 | 95 | 281 | 182 | 0.55 |
| Patient | Age | Survival | ICU | Intubation | COVID-19 | RT-PCR | Male | |
|---|---|---|---|---|---|---|---|---|
| View/Projection | Count | Mean | Y/N | Y/N | Y/N | Count | Positive | Ratio |
| PosteroAnterior (PA) | 326 | 52.21 | 104/19 | 53/67 | 33/55 | 191 | 112 | 0.54 |
| AnteroPosterior (AP) | 192 | 57.93 | 36/13 | 58/6 | 51/15 | 141 | 109 | 0.54 |
| AP Supine | 161 | 57.84 | 51/20 | 89/11 | 42/6 | 136 | 64 | 0.64 |
| Lateral (L) | 82 | 49.78 | 25/2 | 5/11 | 2/16 | 25 | 20 | 0.73 |
3.1 Data Collection
As mentioned earlier, data was largely compiled from public databases on websites such as Radiopaedia.org, the Italian Society of Medical and Interventional Radiology222https://www.sirm.org/category/senza-categoria/covid-19/, Figure1.com333https://www.figure1.com/covid-19-clinical-cases, and the Hannover Medical School [Winther et al., 2020b], both manually and using scrapers. A full list of publications is included in Appendix §C.1.
Images were extracted from online publications, websites, or directly from the PDF using the tool pdfimages444https://poppler.freedesktop.org/. Throughout data collection, we aimed to maintain the quality of the images. Many articles were found using the list of literature provided by Peng et al. [2020]. A limitation of this approach is that we have no control over the processing between the PAC system and the PDF or website. However, we believe that information about the radiological finding was maintained otherwise the image in publication would not be useful. We also believe the images from PDFs and other image sources provide useful information when compared to raw data and expect studies to show this.
A challenge in extracting metadata is the alignment with images. To belong in the same row as an image, a clinical measurement must have been taken on the same day. It can be difficult to automatically determine when the measurement was taken if the metadata appears outside of image captions. For details on scraper design, see §Appendix C. All scripts are made publicly available.
4 Experimental Setup
Using this dataset as a benchmark is very challenging because, by the nature of its construction, it is very biased and unbalanced (recall geographically unbalanced labels in Table 2). This can lead to many negative outcomes if treated as a typical benchmark dataset [Maguolo and Nanni, 2020; Cohen et al., 2020b; Seyyed-Kalantari et al., 2020; Kelly et al., 2019]. Unless otherwise specified only AP and PA views are used to avoid confounding image artifacts of the AP Supine view.
4.1 Leave-One-Country/Continent-Out (LOCO) Evaluation:
In order to deal with issues of bias, we perform a “leave one country/continent out” (LOCO) evaluation. This approach is motivated from training bias common in small, unbalanced datasets. For our evaluation the test set will be composed of data from a single continent. Ideally, we would separate by countries, but this is not possible given the current distribution of data. We also note that not every sample is labelled, so models may be trained and evaluated on data from the same continent, but we ensured that samples in training and evaluation did not originate from the same research group/image source.
This approach should give us a distributional shift that will allow us to correctly evaluate the model. For each task, a subset of the samples will have enough representation to be included. Continents which do not have at least one representative for each class are filtered out and not used.
4.2 Models and Features
In place of images, features are extracted using a pre-trained DenseNet model [Huang et al., 2017] from the TorchXRayVison library [Cohen et al., 2020d] which is trained on 7 large CXR datasets as described in Cohen et al. [2020b]. These datasets were manually aligned with each other on 18 common radiological finding tasks in order to train a model using all datasets at once (atelectasis, consolidation, infiltration, pneumothorax, edema, emphysema, fibrosis, fibrosis, effusion, pneumonia, pleural thickening, cardiomegaly, nodule, mass, hernia, lung lesion, fracture, lung opacity, and enlarged cardiomediastinum). For example “pleural effusion" from one dataset is considered the same as “effusion" from another dataset in order to consider these labels equal. In total, 88,079 non-COVID-19 images were used to train the model on these tasks. We will use the term pre-sigmoid which refers to the output of the last layer of the network multiplied by a weight vector corresponding to each task which would normally then be passed through a sigmoid function. Features will be used in the following constructions (as in Cohen et al. [2020a]):
- •
Intermediate features - the result of the convolutional layers and global averaging (1024 dim vector);
- •
18 outputs - each image was represented by the 18 outputs (pre-sigmoid) here: Atelectasis, Consolidation, Infiltration, Pneumothorax, Edema, Emphysema, Fibrosis, Effusion, Pneumonia, Pleural Thickening, Cardiomegaly, Nodule, Mass, Hernia, Lung Lesion, Fracture, Lung Opacity, Enlarged Cardiomediastinum;
- •
4 outputs - a hand picked subset of the above mentioned outputs (pre-sigmoid) were used containing radiological findings more frequent in pneumonia: Lung Opacity, Pneumonia, Infiltration, and Consolidation;
- •
Lung Opacity output - the single output (pre-sigmoid) for lung opacity was used because it was very related to this task. This feature was different from the opacity score that we would like to predict;
- •
Image pixels - the image itself as a vector of pixels (224224=50176).
- •
Baseline prevalence - to compare against a model which predicted based on the prevalence of the labels only.
Model training is done using linear or logistic regression with default parameters from Sci-kit learn [Pedregosa et al., 2011]. An L2 penalty was used with an LBFGS solver. In the case of “image pixels” an MLP is used with 100 hidden units and ReLU activations Glorot et al. [2011]. It is trained with full batch gradient descent using an Adam optimizer Kingma and Ba [2014] (with , a learning rate of , and 10% of the data used for early stopping.
5 Task Ideas with Baseline Evaluations
We present multiple clinical use cases and potential tools which could be built using this dataset and present a baseline task which is evaluated. We describe the scenarios in detail to both convey what our group has learned while interacting with clinicians as well as solidify what the value is of such a model to avoid misguided efforts solving a problem that doesn’t exist. For the results presented in Tables 5 and 4 a full listing over all data splits is presented in Appendix §C.2.
| Task | Test Regions | Features | # params | AUROC | AUPRC |
|---|---|---|---|---|---|
| COVID-190<offset<8 daysTrue=128False=27 | Americas, Asia, Europe, Oceania | 18 outputs | 18+1 | 0.580.12 | 0.780.16 |
| Intermediate features | 1024+1 | 0.570.12 | 0.770.15 | ||
| 4 outputs | 4+1 | 0.500.01 | 0.730.22 | ||
| Baseline prevalence | 1+1 | 0.500.00 | 0.730.22 | ||
| lung opacity output | 1+1 | 0.500.01 | 0.730.22 | ||
| Image pixels (MLP) | 5017801 | 0.480.06 | 0.720.20 | ||
| Viral or Bacterial0<offset<8 daysTrue=140False=10 | Europe, Oceania | 18 outputs | 18+1 | 0.820.26 | 0.970.04 |
| Intermediate features | 1024+1 | 0.700.19 | 0.840.13 | ||
| lung opacity output | 1+1 | 0.560.08 | 0.720.31 | ||
| 4 outputs | 4+1 | 0.520.03 | 0.710.30 | ||
| Baseline prevalence | 1+1 | 0.500.00 | 0.710.30 | ||
| Image pixels (MLP) | 5017801 | 0.500.01 | 0.710.23 | ||
| Survival prediction0<offset<8 daysTrue=46False=7 | Americas, Asia, Europe | lung opacity output | 1+1 | 0.550.08 | 0.860.05 |
| 4 outputs | 4+1 | 0.540.07 | 0.850.05 | ||
| Baseline prevalence | 1+1 | 0.500.00 | 0.840.04 | ||
| Image pixels (MLP) | 5017801 | 0.500.00 | 0.840.03 | ||
| Intermediate features | 1024+1 | 0.500.00 | 0.840.04 | ||
| 18 outputs | 18+1 | 0.470.03 | 0.840.03 | ||
| ICU stay0<offset<8 daysTrue=17False=27 | Asia, Europe | 18 outputs | 18+1 | 0.810.10 | 0.500.24 |
| Intermediate features | 1024+1 | 0.710.18 | 0.400.21 | ||
| 4 outputs | 4+1 | 0.550.15 | 0.350.36 | ||
| Baseline prevalence | 1+1 | 0.500.00 | 0.280.26 | ||
| lung opacity output | 1+1 | 0.420.12 | 0.280.26 | ||
| Image pixels (MLP) | 5017801 | 0.400.13 | 0.280.20 | ||
| Intubated0<offset<8 daysTrue=12False=23 | Asia, Europe | Intermediate features | 1024+1 | 0.610.06 | 0.400.21 |
| Baseline prevalence | 1+1 | 0.500.00 | 0.320.21 | ||
| Image pixels (MLP) | 5017801 | 0.500.07 | 0.320.17 | ||
| lung opacity output | 1+1 | 0.500.00 | 0.320.21 | ||
| 18 outputs | 18+1 | 0.450.22 | 0.350.26 | ||
| 4 outputs | 4+1 | 0.450.00 | 0.310.20 |
5.1 Complement to COVID-19 Pneumonia Diagnosis and Management
Motivation:
While reverse transcriptase polymerase chain reaction (RTPCR) assay remains the gold standard for diagnosis, CXR plays a major role as the top initial imaging test for patients with suspected COVID-19 pneumonia [Dennie et al., 2020b]. Because of their relative lack of sensitivity (69%, although this was computed with a small sample size) [Wong et al., 2019] and the fact that they are often normal early in the disease [Dennie et al., 2020b; Wong et al., 2019], a negative CXR should not be used to rule out COVID-19 infection [Dennie et al., 2020b]. Instead, features of COVID-19 pneumonia in CXR, although nonspecific, raise the pretest probability of infection. For example, distinguishing between viral and bacterial pneumonia could influence management in addition to other clinical clues [Heneghan et al., 2020]. According to the CSTR and CAR, a CXR is “most useful when an alternative diagnosis is found that completely explains the patient’s presenting symptoms such as, but not limited, to pneumothorax, pulmonary edema, large pleural effusions, lung mass or lung collapse [Dennie et al., 2020b].”
Task Specification:
The hierarchy of labels (Table 3) allows us to perform multiple different classification tasks. A first task is to classify COVID-19 from other causal agents of pneumonia such as bacteria or other viruses. A second task is to distinguish viral from bacterial pneumonia.
Results:
When classifying between COVID/non-COVID most recent works using this dataset have taken other datasets and treated them as non-COVID-19 [Wang and Wong, 2020; Apostolopoulos and Mpesiana, 2020] while results with balanced datasets from Tartaglione et al. [2020] report much lower performance. Our LOCO evaluation aims to avoid these issues and in Table 4 we find that the best performance is only slightly higher than random guessing which would yield a 0.5 AUROC. We specifically note that the “4 outputs” which are commonly associated with Pneumonia are not predictive, possibly implying something about the disease that should be explored more. We find that predicting between bacterial and viral yields reasonably good performance using intermediate features or the 18 specific outputs. This is not surprising as previous work has reported high accuracy when predicting bacterial and viral from pediatric CXR [Kermany et al., 2018].
5.2 Severity Prediction, Including Intensive Care Unit (ICU) Stay
Motivation:
The ICU is reserved for patients who require life support such as mechanical ventilation. In this invasive intervention reserved for patients unable to breathe on their own, an endotracheal tube is inserted into the windpipe (intubation) and the lungs are mechanically inflated and deflated [Tobin and Manthous, 2017]. Predicting the need for mechanical ventilation in advance could help plan management or prepare the patient. Another challenge is knowing when to remove mechanical ventilation (extubation), which falls in a specific window of time [Thille et al., 2013].
Assessing the severity of a patient’s condition using a well adopted scoring system is a key aspect of patient management [Wong et al., 2019; Cohen et al., 2020a; Borghesi and Maroldi, 2020b; Signoroni et al., 2020]. A model which predicts the severity of COVID-19 pneumonia, and pneumonia in general, based on CXR could be used as an assistive tool when managing patient care for escalation or de-escalation of care, especially in the ICU.
Non-ML work by Allenbach et al. [2020] combines information from CT scans and CXR with other clinical information to create a score-based predictive model for transfer to the ICU.
This task is further motivated by papers published after a preprint of this work which employed siamese networks to represent severity [Li et al., 2020] as well as predicted deterioration risk scores and time estimates [Shamout et al., 2020].
It is important to note with this task that predicting these events could be confounded by the presence of an endotracheal tube in the CXR of a mechanically ventilated patient; when available, this is annotated in our dataset.
Due to the reduced mobility of ICU patients, CXR are often obtained with the patient lying down in a view referred to as “AP supine” [Khan et al., 2009], which includes but is not limited to patients who are intubated or soon to be. Because this position drastically modifies the appearance of the CXR, a naive approach could confound such changes with the need to be intubated.
Task Specification:
The first thing that is required for this task is a well defined scoring system that defines patient severity. The mRALE (modified Radiographic Assessment of Lung Edema) score [Wong et al., 2019; Cohen et al., 2020a] was developed specifically with in context of assessing COVID-19 severity from CXR and was used to score 94 images in this dataset in Cohen et al. [2020a] by two chest radiologists and a radiology resident. Also, the Brixia score [Borghesi and Maroldi, 2020b; Signoroni et al., 2020] was used by another group to score 192 of these images by "two expert radiologists, a board-certified staff member and a trainee with 22 and 2 years of experience respectively" [Signoroni et al., 2020].
Our task and evaluation will use the severity scores created by Cohen et al. [2020a]. This work provides two scores: Geographic Extent (0-8), how much opacity covers the lungs, and Opacity (0-6), how opaque the lungs are.
Also, an ICU stay and the patient being intubated are both predicted given patients between 0 and 8 days from symptoms/presentation. Images marked as intubation present are excluded from the evaluation as this would be a visible confounder in the image. For ICU stay prediction images marked as already in the ICU are excluded.
Results:
Table 5 shows performance similar to that reported by Cohen et al. [2020a]. The evaluation used in that work is not LOCO.
Table 4 shows ICU stay is predicted reasonably well at 0.81 AUROC using all 18 outputs. The improvement over the “4 outputs” which are associated with Pneumonia may imply that ICU stay is predictive by features not related to pneumonia. Intubation is predicted poorly.
| Task | Test Regions | Features |
|
| MAE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| GeographicExtentScore (0-8)N=94 | Asia, Europe | 4 outputs | 4+1 | 0.820.05 | 0.620.05 | 1.110.08 | ||||
| 18 outputs | 18+1 | 0.800.09 | 0.590.17 | 1.150.15 | ||||||
| lung opacity output | 1+1 | 0.800.05 | 0.580.02 | 1.150.01 | ||||||
| Intermediate features | 1024+1 | 0.770.06 | 0.410.17 | 1.410.08 | ||||||
| Baseline prevalence | 1+1 | 0.000.00 | -0.330.25 | 2.140.31 | ||||||
| OpacityScore (0-6)N=94 | Asia, Europe | 4 outputs | 4+1 | 0.790.07 | 0.610.11 | 0.730.10 | ||||
| lung opacity output | 1+1 | 0.790.07 | 0.600.09 | 0.760.10 | ||||||
| 18 outputs | 18+1 | 0.660.13 | 0.290.26 | 0.900.16 | ||||||
| Intermediate features | 1024+1 | 0.680.11 | -0.090.45 | 1.200.26 | ||||||
| Baseline prevalence | 1+1 | 0.000.00 | -0.260.20 | 1.300.00 |
5.3 Survival outcome
Motivation:
Not too dissimilar to severity prediction, determining at what point survival can be predicted could be useful for patient management. Given a series of patient chest X-rays over time, it could be possible to determine the probability of survival.
Non-ML models are able to predict in-hospital mortality for patients with COVID-19 using an original severity score for CXR (Brixia score) combined with two predictive factors, which are patient age and immunosuppressive conditions [Borghesi et al., 2020a]. ML models are able to predict survival based on clinical features (lactate dehydrogenase, lymphocyte count, and high-sensitivity C-reactive protein) with high accuracy [Yan et al., 2020].
Task Specification:
In order to make predictions which are relevant to the clinical context, it is important to control for the time period when the patient is observed. Our evaluation is on data between 0 and 8 days since symptoms or admission in order to simulate predicting at the beginning of management. Predictions will be made on non-intubated patients to avoid this confounder of severity.
Results:
In Table 4, almost random performance of 0.55 AUROC is obtained using lung opacity and the 4 hand picked features of Pneumonia. In general, no method works well.
5.4 Trajectory Prediction
Motivation:
The ability to gauge severity of COVID-19 lung infections could be used to complement other severity tools for escalation or de-escalation of care, especially in the ICU. Following diagnosis, patients’ CXR could be scored periodically to objectively and quantitatively track pulmonary disease progression and treatment response. Eventually, physicians could track patients’ response to various drugs and treatments using CXR and uploading the images to the dataset, allowing researchers to create predictive tools to measure recuperation. Such a model could also be used as an objective tool to compare response to different management algorithms and inspire better management strategies.
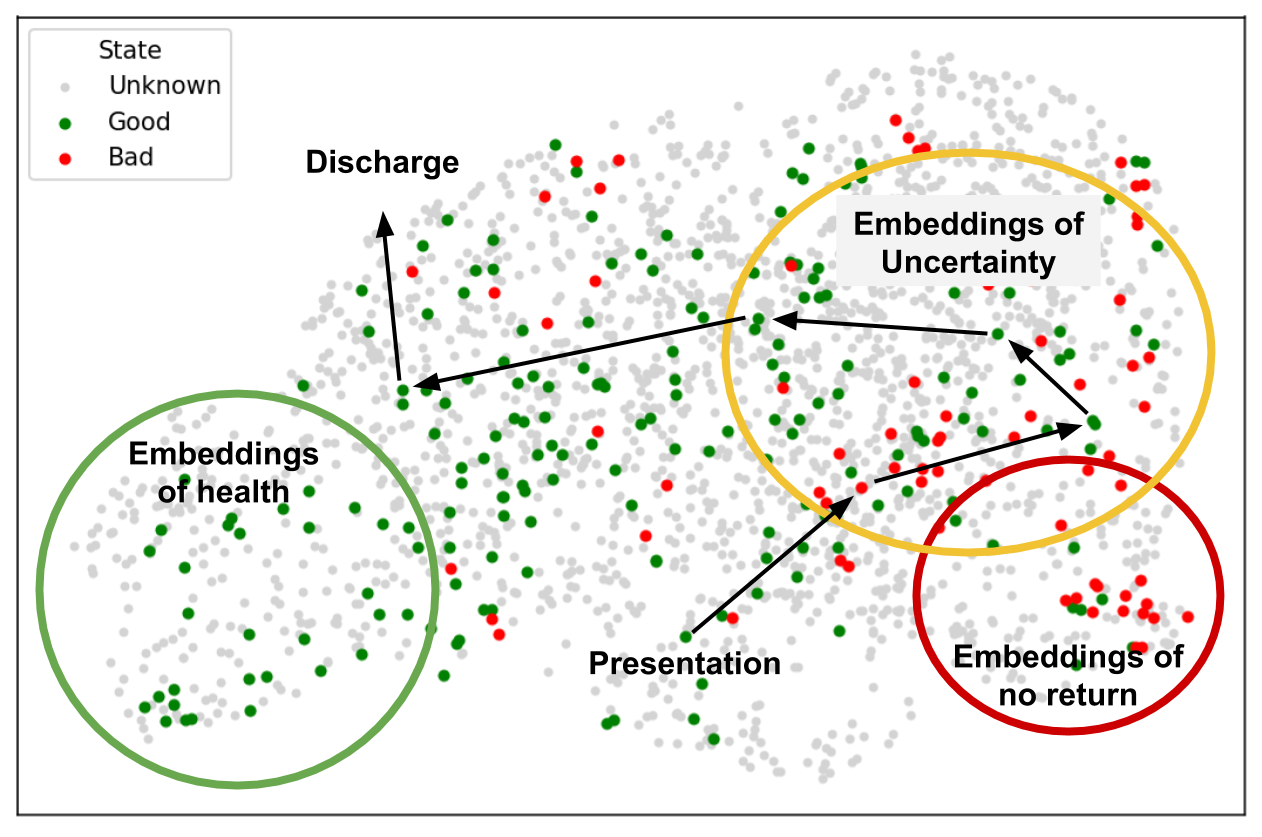
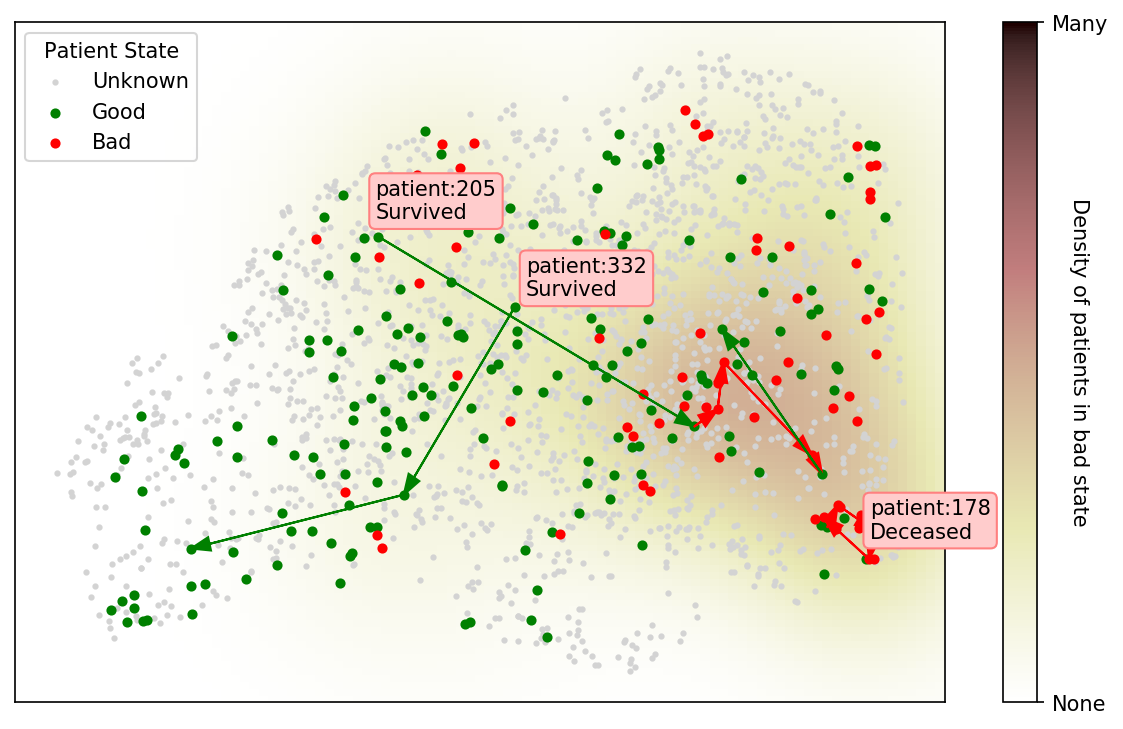
If the representation is expressive enough, patients can be plotted as shown in Figure 4a. A conceptual figure and our current realization are shown using a pre-trained CXR model showing the available trajectories and patient outcomes. This approach could serve as a way to iterate quickly with a medical team (to simply explore the learned representation instead of building complete tools) to make sense of the complexities of these models and patients.
Task Specification:
Using this dataset, we could visualize a model’s representation of CXRs and plot the trajectory of patients according to a color scheme representing their state/image (good or bad). The representation is based off of the 18 pre-sigmoid outputs of the DenseNet discussed. A good state is defined as non-intubated, not in the ICU, or the last state before discharge. A bad state is defined as intubated, in the ICU, or any state which is the last in the series and the patient is determined to have died. A kernel density estimation is taken of the 2d embeddings of all bad states to illustrate severity. Here PA, AP, and AP Supine images are used to maximize the data visualized as we did not observe any shift in the representation.
Results:
Figure 4b shows proximity of image embeddings which represent patients in a good or bad state, demonstrating the potential insight already contained in these pre-trained models. The three patients display trajectories as we would imagine. Patient 178555https://www.eurorad.org/case/16660 spirals in an area which appears bad. Patient 205666https://radiopaedia.org/cases/covid-19-pneumonia-progression-and-regression progresses into bad states but manages to pull themselves out. Patient 332 [Sivakorn et al., 2020] seems to recover quickly to a region which is only populated by good states.
6 Future Task Ideas
There are many potential tasks that we were not able to explore in this work, primarily the use of saliency maps and the utility of out-of-distribution models.
Evaluating saliency maps can be challenging, but it is a useful evaluation for methods which aim to explain model predictions [Taghanaki et al., 2019; Singla et al., 2020]. Our dataset contains lung bounding boxes, which were contributed by Andrew Gough at General Blockchain Inc. for 167 images annotated for the left and right lung. Also, 209 generated lung segmentations were added to the dataset by Selvan et al. [2020]; the team trained a model on an external dataset and applied it to our dataset. As there are no “ground truth” segmentations, these can be used to examine if saliency maps are located reasonably within lung regions to detect overfitting. This can be seen by checking if the predictive regions of the image lie outside of the region of interest [Ross et al., 2017; Badgeley et al., 2019; Viviano et al., 2019].
General anomaly detection could be a useful model when trying to identify what is new about an illness such as COVID-19. Out-of-distribution (OoD) tools [Shafaei et al., 2018] or unpaired distribution matching models [Zhu et al., 2017; Kim et al., 2017] could capture the shift in distributions and present them as changes to images Cohen et al. [2018]. Identifying what about the COVID-19 distribution is different from other viral or bacterial pneumonias could aid in studying both the disease as well as the models representations for overfitting [Singla et al., 2020]. Transfer learning methods actively under development in the ML community such as few/zero-shot [Wang et al., 2019; Tian et al., 2020b; Larochelle et al., 2008; Ren et al., 2019], meta-learning [Andrychowicz et al., 2016; Snell et al., 2017], deep metric learning [Roth et al., 2020], and domain adaptation [Motiian et al., 2017] will likely be useful in this setting.
7 Conclusion
This paper presents a dataset of COVID-19 images together with clinical metadata which can be used for a variety of tasks. This dataset puts existing ML algorithms to the test. Given the number of existing large CXR datasets, novel tasks related to COVID-19 present a relevant challenge to overcome. We note that a major limitation of this work is the selection bias when gathering publicly available images which are likely made public for educational reasons because they are clear examples or interesting cases. Therefore, they do not represent the real world distribution of cases. Furthermore, another selection bias is that the information given on public platforms such as Figure 1 or Radiopaedia might not be complete for all patients and/or omit normal values (e.g., presence or absence of transfer to the ICU, lymphocyte count). Lastly, we do not yet have variables such as ethnic background, pre-existing conditions, and immunosuppression status. Any clinical claims made from models must therefore be backed by rigorous evaluation and take into account these limitations. Nevertheless, we believe that this dataset and the discussion of clinical context will contribute towards the machine learning community developing solutions with potential use in healthcare.
Broader Impact
This project aims to make a dataset of patients with a novel life-threatening disease accessible to researchers so that tools can be created to aid in the care of future patients. The manner in which we collect existing public data ensures that patients are not put at risk.
Data impact: Image data linked with clinically relevant attributes in a public dataset that is designed for ML will enable parallel development of diagnosis and management tools and rapid local validation of models. Furthermore, this data can be used for a variety of different tasks.
Tool impact: Tools developed using this data and with the ideas presented can give physicians an edge and allow them to act with more confidence while they wait for the analysis of a radiologist by having a digital second opinion confirm their assessment of a patient’s condition. In addition, these tools can provide quantitative scores which can enable large scale analysis of CXR without the need for costly/time consuming manual annotations.
Acknowledgements
We thank Dr. Errol Colak, Luke Oakden-Rayner, Rupert Brooks, Hadrien Bertrand, Dr. Michaël Chassé, and Dr. Carl Chartrand-Lefebvre for their input. This research is based on work partially supported by the CIFAR AI and COVID-19 Catalyst Grants. This work utilized the supercomputing facilities managed by Compute Canada and Calcul Quebec. We thank AcademicTorrents.com for making data available for our research.
Ethical Standards
The work follows appropriate ethical standards in conducting research and writing the manuscript, following all applicable laws and regulations regarding treatment of animals or human subjects. This project is approved by the University of Montreal’s Ethics Committee #CERSES-20-058-D
Conflicts of Interest
We declare we don’t have conflicts of interest.
References
- Ackermann et al. [2020] Maximilian Ackermann, Stijn E Verleden, Mark Kuehnel, Axel Haverich, Tobias Welte, Florian Laenger, Arno Vanstapel, Christopher Werlein, Helge Stark, Alexandar Tzankov, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. New England Journal of Medicine, 2020.
- ACR [2018] ACR. ACR Appropriateness Criteria for Acute Respiratory Illness in Immunocompetent Patients. Technical report, American College of Radiology, 2018.
- ACR [2020] ACR. ACR Recommendations for the use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection. Technical report, American College of Radiology, 2020. URL https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection.
- Ahmed et al. [2020] Taha Ahmed, Ronak J Shah, Shab E Gul Rahim, Monica Flores, and Amy OLinn. Coronavirus disease 2019 (covid-19) complicated by acute respiratory distress syndrome: An internist’s perspective. Cureus, 2020. doi: 10.7759/cureus.7482.
- Aigner et al. [2020] Clemens Aigner, Ulf Dittmer, Markus Kamler, Stephane Collaud, and Christian Taube. Covid-19 in a lung transplant recipient. The Journal of Heart and Lung Transplantation, 2020. doi: 10.1016/j.healun.2020.04.004.
- Allen et al. [2010] Carolyn M. Allen, Hamdan H. Al-Jahdali, Klaus L. Irion, Sarah Ai Al Ghanem, Alaa Gouda, and Ali Nawaz Khan. Imaging lung manifestations of HIV/AIDS, 10 2010. ISSN 18171737.
- Allenbach et al. [2020] Yves Allenbach, David Saadoun, Georgina Maalouf, Matheus Vieira, Alexandra Hellio, Jacques Boddaert, Helene Gros, Joe Elie Salem, Matthieu Resche-Rigon, Lucie Biard, Olivier Benveniste, and Patrice Cacoub. Multivariable prediction model of intensive care unit transfer and death: a French prospective cohort study of COVID-19 patients. medRxiv, page 2020.05.04.20090118, 5 2020. doi: 10.1101/2020.05.04.20090118.
- Amer et al. [2020] Rula Amer, Maayan Frid-Adar, Ophir Gozes, Jannette Nassar, and Hayit Greenspan. COVID-19 in CXR: from Detection and Severity Scoring to Patient Disease Monitoring. 8 2020. URL http://arxiv.org/abs/2008.02150.
- An et al. [2020] Peng An, Ping Song, Kai Lian, and Yong Wang. Ct manifestations of novel coronavirus pneumonia: A case report. Balkan Medical Journal, 2020. doi: 10.4274/balkanmedj.galenos.2020.2020.2.15.
- Andrychowicz et al. [2016] Marcin Andrychowicz, Misha Denil, Sergio Gómez Colmenarejo, Matthew W. Hoffman, David Pfau, Tom Schaul, Brendan Shillingford, Nando de Freitas, Sergio Gomez, Matthew W. Hoffman, David Pfau, Tom Schaul, and Nando de Freitas. Learning to learn by gradient descent by gradient descent. Neural Information Processing Systems, 6 2016. ISSN 0219-1377. doi: 10.1007/s10115-008-0151-5. URL http://arxiv.org/abs/1606.04474.
- Apostolopoulos and Mpesiana [2020] Ioannis D. Apostolopoulos and Tzani A. Mpesiana. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, pages 1–6, 4 2020. ISSN 2662-4729. doi: 10.1007/s13246-020-00865-4.
- Avula et al. [2020] Akshay Avula, Krishna Nalleballe, Naureen Narula, Steven Sapozhnikov, Vasuki Dandu, Sudhamshi Toom, Allison Glaser, and Dany Elsayegh. Covid-19 presenting as stroke. Brain, Behavior, and Immunity, 2020. doi: 10.1016/j.bbi.2020.04.077.
- Azekawa et al. [2020] Shuhei Azekawa, Ho Namkoong, Keiko Mitamura, Yoshihiro Kawaoka, and Fumitake Saito. Co-infection with SARS-CoV-2 and influenza a virus. IDCases, 20:e00775, 2020. doi: 10.1016/j.idcr.2020.e00775. URL https://doi.org/10.1016/j.idcr.2020.e00775.
- Badgeley et al. [2019] Marcus A. Badgeley, John R. Zech, Luke Oakden-Rayner, Benjamin S. Glicksberg, Manway Liu, William Gale, Michael V. McConnell, Bethany Percha, Thomas M. Snyder, and Joel T. Dudley. Deep learning predicts hip fracture using confounding patient and healthcare variables. npj Digital Medicine, 2(1):1–10, 12 2019. ISSN 2398-6352. doi: 10.1038/s41746-019-0105-1.
- Ball et al. [2017] Lorenzo Ball, Veronica Vercesi, Federico Costantino, Karthikka Chandrapatham, and Paolo Pelosi. Lung imaging: How to get better look inside the lung, 7 2017. ISSN 23055847.
- Banerjee et al. [2020] Debasish Banerjee, Joyce Popoola, Sapna Shah, Irina Chis Ster, Virginia Quan, and Mysore Phanish. Covid-19 infection in kidney transplant recipients. Kidney International, 2020. doi: 10.1016/j.kint.2020.03.018.
- Beek [2015] Edwin JR van Beek. Lung cancer screening: Computed tomography or chest radiographs? World Journal of Radiology, 7(8):189, 2015. ISSN 1949-8470. doi: 10.4329/wjr.v7.i8.189.
- Bhatraju et al. [2020] Pavan K. Bhatraju, Bijan J. Ghassemieh, Michelle Nichols, Richard Kim, Keith R. Jerome, Arun K. Nalla, Alexander L. Greninger, Sudhakar Pipavath, Mark M. Wurfel, Laura Evans, Patricia A. Kritek, T. Eoin West, Andrew Luks, Anthony Gerbino, Chris R. Dale, Jason D. Goldman, Shane O’Mahony, and Carmen Mikacenic. Covid-19 in critically ill patients in the seattle region —case series. New England Journal of Medicine, 2020. doi: 10.1056/nejmoa2004500.
- Borghesi and Maroldi [2020a] Andrea Borghesi and Roberto Maroldi. Covid-19 outbreak in italy: experimental chest x-ray scoring system for quantifying and monitoring disease progression. La radiologia medica, 2020a. doi: 10.1007/s11547-020-01200-3.
- Borghesi and Maroldi [2020b] Andrea Borghesi and Roberto Maroldi. COVID-19 outbreak in Italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. Radiologia Medica, 2020b. ISSN 18266983. doi: 10.1007/s11547-020-01200-3.
- Borghesi et al. [2020a] Andrea Borghesi, Angelo Zigliani, Salvatore Golemi, Nicola Carapella, Patrizia Maculotti, Davide Farina, and Roberto Maroldi. Chest X-ray severity index as a predictor of in-hospital mortality in coronavirus disease 2019: A study of 302 patients from Italy. International Journal of Infectious Diseases, 5 2020a. ISSN 12019712. doi: 10.1016/j.ijid.2020.05.021.
- Borghesi et al. [2020b] Andrea Borghesi, Angelo Zigliani, Roberto Masciullo, Salvatore Golemi, Patrizia Maculotti, Davide Farina, and Roberto Maroldi. Radiographic severity index in covid-19 pneumonia: relationship to age and sex in 783 italian patients. La radiologia medica, 2020b. doi: 10.1007/s11547-020-01202-1.
- Bustos et al. [2019] Aurelia Bustos, Antonio Pertusa, Jose-Maria Salinas, and Maria de la Iglesia-Vayá. PadChest: A large chest x-ray image dataset with multi-label annotated reports. arXiv preprint, 1 2019. URL http://arxiv.org/abs/1901.07441.
- Cai et al. [2020] Xiao Qing Cai, Pi Qi Jiao, Tao Wu, Fu Ming Chen, Bao Shi Han, Jiu Cong Zhang, Yong Jiu Xiao, Zhi Feng Chen, Jun Li, Yu Ying Zhao, Ling Ma, Yan Liu, Ya Jun Shi, Pei Jun Dai, and Yun Dai Chen. Armarium facilitating angina management post myocardial infarction concomitant with coronavirus disease 2019, 2020. ISSN 16715411.
- Chen et al. [2020] Nanshan Chen, Min Zhou, Xuan Dong, Jieming Qu, Fengyun Gong, Yang Han, Yang Qiu, Jingli Wang, Ying Liu, Yuan Wei, Jiaan Xia, Ting Yu, Xinxin Zhang, and Li Zhang. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, china: a descriptive study. The Lancet, 395(10223):507–513, February 2020. doi: 10.1016/s0140-6736(20)30211-7. URL https://doi.org/10.1016/s0140-6736(20)30211-7.
- Cheng et al. [2020] Shao-Chung Cheng, Yuan-Chia Chang, Yu-Long Fan Chiang, Yu-Chan Chien, Mingte Cheng, Chin-Hua Yang, Chia-Husn Huang, and Yuan-Nian Hsu. First case of coronavirus disease 2019 (COVID-19) pneumonia in taiwan. Journal of the Formosan Medical Association, 119(3):747–751, March 2020. doi: 10.1016/j.jfma.2020.02.007. URL https://doi.org/10.1016/j.jfma.2020.02.007.
- Cohen et al. [2018] Joseph Paul Cohen, Margaux Luck, and Sina Honari. Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. In Medical Image Computing & Computer Assisted Intervention (MICCAI), 2018.
- Cohen et al. [2020a] Joseph Paul Cohen, Lan Dao, Paul Morrison, Karsten Roth, Yoshua Bengio, Beiyi Shen, Almas Abbasi, Mahsa Hoshmand-Kochi, Marzyeh Ghassemi, Haifang Li, and Tim Q Duong. Predicting COVID-19 Pneumonia Severity on Chest X-ray with Deep Learning. arXiv:2005.11856, 5 2020a. URL http://arxiv.org/abs/2005.11856.
- Cohen et al. [2020b] Joseph Paul Cohen, Mohammad Hashir, Rupert Brooks, and Hadrien Bertrand. On the limits of cross-domain generalization in automated X-ray prediction. In Medical Imaging with Deep Learning, 2020b. URL https://arxiv.org/abs/2002.02497.
- Cohen et al. [2020c] Joseph Paul Cohen, Paul Morrison, and Lan Dao. COVID-19 Image Data Collection. https://github.com/ieee8023/covid-chestxray-dataset, 2020c. URL https://arxiv.org/abs/2003.11597.
- Cohen et al. [2020d] Joseph Paul Cohen, Joseph Viviano, Mohammad Hashir, and Hadrien Bertrand. TorchXRayVision: A library of chest X-ray datasets and models. https://github.com/mlmed/torchxrayvision, 2020d. URL https://github.com/mlmed/torchxrayvision.
- Coimbra et al. [2020] Raul Coimbra, Sara Edwards, Hayato Kurihara, Gary Alan Bass, Zsolt J. Balogh, Jonathan Tilsed, Roberto Faccincani, Michele Carlucci, Isidro Martínez Casas, Christine Gaarder, Arnold Tabuenca, Bruno C. Coimbra, and Ingo Marzi. European society of trauma and emergency surgery (estes) recommendations for trauma and emergency surgery preparation during times of covid-19 infection. European Journal of Trauma and Emergency Surgery, 2020. doi: 10.1007/s00068-020-01364-7.
- Couzin-Frankel [2019] Jennifer Couzin-Frankel. Medicine contends with how to use artificial intelligence, 6 2019. ISSN 10959203. URL https://science.sciencemag.org/content/364/6446/1119https://science.sciencemag.org/content/364/6446/1119.abstract.
- Cuong et al. [2020] Le Van Cuong, Hoang Thi Nam Giang, Le Khac Linh, Jaffer Shah, Le Van Sy, Trinh Huu Hung, Abdullah Reda, Luong Ngoc Truong, Do Xuan Tien, and Nguyen Tien Huy. The first vietnamese case of COVID-19 acquired from china. The Lancet Infectious Diseases, February 2020. doi: 10.1016/s1473-3099(20)30111-0. URL https://doi.org/10.1016/s1473-3099(20)30111-0.
- Dastan et al. [2020] Farzaneh Dastan, Ali Saffaei, Seyed Mehdi Mortazavi, Hamidreza Jamaati, Nadia Adnani, Sasan Samiee Roudi, Arda Kiani, Atefeh Abedini, and Seyed MohammadReza Hashemian. Continues renal replacement therapy (crrt) with disposable hemoperfusion cartridge: A promising option for severe covid-19. Journal of Global Antimicrobial Resistance, 2020. doi: 10.1016/j.jgar.2020.04.024.
- De La Iglesia Vayá et al. [2020] Maria De La Iglesia Vayá, Jose Manuel Saborit, Joaquim Angel Montell, Antonio Pertusa, Aurelia Bustos, Miguel Cazorla, Joaquin Galant, Xavier Barber, Domingo Orozco-Beltrán, Francisco Garcia, Marisa Caparrós, Germán González, and Jose María Salinas. BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. arXiv:2006.01174, 2020.
- DeGrave et al. [2020] Alex J DeGrave, Joseph D Janizek, and Su-In Lee. AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv, page 2020.09.13.20193565, 1 2020. doi: 10.1101/2020.09.13.20193565. URL http://medrxiv.org/content/early/2020/09/14/2020.09.13.20193565.abstract.
- Demner-Fushman et al. [2016] Dina Demner-Fushman, Marc D. Kohli, Marc B. Rosenman, Sonya E. Shooshan, Laritza Rodriguez, Sameer Antani, George R. Thoma, and Clement J. McDonald. Preparing a collection of radiology examinations for distribution and retrieval. Journal of the American Medical Informatics Association, 23(2):304–310, 3 2016. ISSN 1527974X. doi: 10.1093/jamia/ocv080.
- Dennie et al. [2020a] Carole Dennie, Cameron Hague, Robert S. Lim, Daria Manos, Brett F. Memauri, Elsie T. Nguyen, and Jana Taylor. Canadian Society of Thoracic Radiology/Canadian Association of Radiologists Consensus Statement Regarding Chest Imaging in Suspected and Confirmed COVID-19, 2020a. ISSN 14882361. URL http://journals.sagepub.com/doi/10.1177/0846537120924606.
- Dennie et al. [2020b] Carole Dennie, Cameron Hague, Robert S Lim, Daria Manos, Brett F Memauri, Elsie T Nguyen, and Jana Taylor. The Canadian Society of Thoracic Radiology (CSTR) and Canadian Association of Radiologists (CAR) Consensus Statement Regarding Chest Imaging in Suspected and Confirmed COVID-19. Technical report, 4 2020b. URL https://car.ca/covid-19/.
- Ebrille et al. [2020] Elisa Ebrille, Maria Teresa Lucciola, Claudia Amellone, Flavia Ballocca, Fabrizio Orlando, and Massimo Giammaria. Syncope as the presenting symptom of covid-19 infection. HeartRhythm Case Reports, 2020. doi: 10.1016/j.hrcr.2020.04.015.
- FDA [2017] FDA. What are the Radiation Risks from CT? Technical report, U.S. Food and Drug Administration, 2017. URL https://www.fda.gov/radiation-emitting-products/medical-x-ray-imaging/what-are-radiation-risks-ct.
- Fichera et al. [2020] Giulia Fichera, Roberto Stramare, Giorgio De Conti, Raffaella Motta, and Chiara Giraudo. It’s not over until it’s over: the chameleonic behavior of covid-19 over a six-day period. La radiologia medica, 2020. doi: 10.1007/s11547-020-01203-0.
- Fidan [2020] Vural Fidan. New type of corona virus induced acute otitis media in adult. American Journal of Otolaryngology, 2020. doi: 10.1016/j.amjoto.2020.102487.
- Filatov et al. [2020] Asia Filatov, Pamraj Sharma, Fawzi Hindi, and Patricio S Espinosa. Neurological complications of coronavirus disease (covid-19): Encephalopathy. Cureus, 2020. doi: 10.7759/cureus.7352.
- Floca [2014] Ralf Floca. Challenges of Open Data in Medical Research. In Opening Science, pages 297–307. Springer International Publishing, 2014. doi: 10.1007/978-3-319-00026-8{\_}22.
- Glorot et al. [2011] Xavier Glorot, Antoine Bordes, and Yoshua Bengio. Deep sparse rectifier neural networks. In Journal of Machine Learning Research, volume 15, pages 315–323. Journal of Machine Learning Research, 6 2011. URL http://proceedings.mlr.press/v15/glorot11a.html.
- Gordienko et al. [2018] Yu. Gordienko, Peng Gang, Jiang Hui, Wei Zeng, Yu. Kochura, O Alienin, O Rokovyi, and S Stirenko. Deep Learning with Lung Segmentation and Bone Shadow Exclusion Techniques for Chest X-Ray Analysis of Lung Cancer. Advances in Computer Science for Engineering and Education, page 638–647, 5 2018. ISSN 2194-5365. doi: 10.1007/978-3-319-91008-6{\_}63. URL http://dx.doi.org/10.1007/978-3-319-91008-6_63.
- Hao [2020] Karen Hao. AI is helping triage coronavirus patients. The tools may be here to stay. MIT Technology Review, 2020. URL https://www.technologyreview.com/2020/04/23/1000410/ai-triage-covid-19-patients-health-care/.
- Hare et al. [2020] S.S. Hare, J.C.L. Rodrigues, A. Nair, J. Jacob, S. Upile, A. Johnstone, R. Mcstay, A. Edey, and G. Robinson. The continuing evolution of covid-19 imaging pathways in the uk: a british society of thoracic imaging expert reference group update. Clinical Radiology, 2020. doi: 10.1016/j.crad.2020.04.002.
- Hashir et al. [2020] Mohammad Hashir, Hadrien Bertrand, and Joseph Paul Cohen. Quantifying the Value of Lateral Views in Deep Learning for Chest X-rays. In Medical Imaging with Deep Learning, 2020.
- Heneghan et al. [2020] Carl Heneghan, Annette Pluddemann, and Kamal R. Mahtani. Differentiating viral from bacterial pneumonia. Technical report, Centre for Evidence-Based Medicine, Nuffield Department of Primary Care Health Sciences University of Oxford, 2020. URL https://www.cebm.net/covid-19/differentiating-viral-from-bacterial-pneumonia/.
- Hiramatsu et al. [2020] Mariko Hiramatsu, Naoki Nishio, Masayuki Ozaki, Yuichiro Shindo, Katsunao Suzuki, Takanori Yamamoto, Yasushi Fujimoto, and Michihiko Sone. Anesthetic and surgical management of tracheostomy in a patient with covid-19. Auris Nasus Larynx, 2020. doi: 10.1016/j.anl.2020.04.002.
- Holshue et al. [2020] Michelle L. Holshue, Chas DeBolt, Scott Lindquist, Kathy H. Lofy, John Wiesman, Hollianne Bruce, Christopher Spitters, Keith Ericson, Sara Wilkerson, Ahmet Tural, George Diaz, Amanda Cohn, LeAnne Fox, Anita Patel, Susan I. Gerber, Lindsay Kim, Suxiang Tong, Xiaoyan Lu, Steve Lindstrom, Mark A. Pallansch, William C. Weldon, Holly M. Biggs, Timothy M. Uyeki, and Satish K. Pillai. First case of 2019 novel coronavirus in the united states. New England Journal of Medicine, 382(10):929–936, March 2020. doi: 10.1056/nejmoa2001191. URL https://doi.org/10.1056/nejmoa2001191.
- Hsih et al. [2020] Wen-Hsin Hsih, Meng-Yu Cheng, Mao-Wang Ho, Chia-Huei Chou, Po-Chang Lin, Chih-Yu Chi, Wei-Chih Liao, Chih-Yu Chen, Lih-Ying Leong, Ni Tien, Huan-Cheng Lai, Yi-Chyi Lai, and Min-Chi Lu. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central taiwan. Journal of Microbiology, Immunology and Infection, March 2020. doi: 10.1016/j.jmii.2020.03.008. URL https://doi.org/10.1016/j.jmii.2020.03.008.
- Huang et al. [2017] Gao Huang, Zhuang Liu, Laurens van der Maaten, and Kilian Q. Weinberger. Densely Connected Convolutional Networks. In Computer Vision and Pattern Recognition, 2017. URL https://arxiv.org/abs/1608.06993.
- Huang et al. [2020] Wei-Hsuan Huang, Ling-Chiao Teng, Ting-Kuang Yeh, Yu-Jen Chen, Wei-Jung Lo, Ming-Ju Wu, Chun-Shih Chin, Yu-Tse Tsan, Tzu-Chieh Lin, Jyh-Wen Chai, Chin-Fu Lin, Chien-Hao Tseng, Chia-Wei Liu, Chi-Mei Wu, Po-Yen Chen, Zhi-Yuan Shi, and Po-Yu Liu. 2019 novel coronavirus disease (covid-19) in taiwan: Reports of two cases from wuhan, china. Journal of Microbiology, Immunology and Infection, 2020. doi: 10.1016/j.jmii.2020.02.009.
- Irvin et al. [2019] Jeremy Irvin, Pranav Rajpurkar, Michael Ko, Yifan Yu, Silviana Ciurea-Ilcus, Chris Chute, Henrik Marklund, Behzad Haghgoo, Robyn Ball, Katie Shpanskaya, Jayne Seekins, David A. Mong, Safwan S. Halabi, Jesse K. Sandberg, Ricky Jones, David B. Larson, Curtis P. Langlotz, Bhavik N. Patel, Matthew P. Lungren, and Andrew Y. Ng. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In AAAI Conference on Artificial Intelligence, 1 2019. URL http://arxiv.org/abs/1901.07031.
- Islam and Zhang [2018] Jyoti Islam and Yanqing Zhang. Towards robust lung segmentation in chest radiographs with deep learning, 2018.
- Islam et al. [2017] Mohammad Tariqul Islam, Md Abdul Aowal, Ahmed Tahseen Minhaz, and Khalid Ashraf. Abnormality detection and localization in chest x-rays using deep convolutional neural networks, 2017.
- Jin et al. [2020] Ying-Hui Jin, , Lin Cai, Zhen-Shun Cheng, Hong Cheng, Tong Deng, Yi-Pin Fan, Cheng Fang, Di Huang, Lu-Qi Huang, Qiao Huang, Yong Han, Bo Hu, Fen Hu, Bing-Hui Li, Yi-Rong Li, Ke Liang, Li-Kai Lin, Li-Sha Luo, Jing Ma, Lin-Lu Ma, Zhi-Yong Peng, Yun-Bao Pan, Zhen-Yu Pan, Xue-Qun Ren, Hui-Min Sun, Ying Wang, Yun-Yun Wang, Hong Weng, Chao-Jie Wei, Dong-Fang Wu, Jian Xia, Yong Xiong, Hai-Bo Xu, Xiao-Mei Yao, Yu-Feng Yuan, Tai-Sheng Ye, Xiao-Chun Zhang, Ying-Wen Zhang, Yin-Gao Zhang, Hua-Min Zhang, Yan Zhao, Ming-Juan Zhao, Hao Zi, Xian-Tao Zeng, Yong-Yan Wang, and Xing-Huan Wang. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-ncov) infected pneumonia (standard version). Military Medical Research, 2020. doi: 10.1186/s40779-020-0233-6.
- jin Zhang et al. [2020] Jin jin Zhang, Xiang Dong, Yi yuan Cao, Ya dong Yuan, Yi bin Yang, You qin Yan, Cezmi A. Akdis, and Ya dong Gao. Clinical characteristics of 140 patients infected with SARS-CoV-2 in wuhan, china. Allergy, February 2020. doi: 10.1111/all.14238. URL https://doi.org/10.1111/all.14238.
- Johnson et al. [2019] Alistair E. W. Johnson, Tom J. Pollard, Seth J. Berkowitz, Nathaniel R. Greenbaum, Matthew P. Lungren, Chih-ying Deng, Roger G. Mark, and Steven Horng. MIMIC-CXR: A large publicly available database of labeled chest radiographs. Nature Scientific Data, 1 2019. doi: 10.1038/s41597-019-0322-0. URL http://arxiv.org/abs/1901.07042.
- Kanne et al. [2020] Jeffrey P. Kanne, Brent P. Little, Jonathan H. Chung, Brett M. Elicker, and Loren H. Ketai. Essentials for Radiologists on COVID-19: An Update-Radiology Scientific Expert Panel. Radiology, 2020. ISSN 15271315. doi: 10.1148/radiol.2020200527.
- Keen et al. [2013] Justin Keen, Radu Calinescu, Richard Paige, and John Rooksby. Big data {+} politics = open data: The case of health care data in England. Policy and Internet, 5(2):228–243, 6 2013. ISSN 19442866. doi: 10.1002/1944-2866.POI330. URL http://doi.wiley.com/10.1002/1944-2866.POI330.
- Kelly et al. [2019] Christopher J. Kelly, Alan Karthikesalingam, Mustafa Suleyman, Greg Corrado, and Dominic King. Key challenges for delivering clinical impact with artificial intelligence, 10 2019. ISSN 17417015. URL https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1426-2.
- Kermany et al. [2018] Daniel S Kermany, Michael Goldbaum, Wenjia Cai, Carolina C S Valentim, Huiying Liang, Sally L Baxter, Alex McKeown, Ge Yang, Xiaokang Wu, Fangbing Yan, Justin Dong, Made K Prasadha, Jacqueline Pei, Magdalene Y L Ting, Jie Zhu, Christina Li, Sierra Hewett, Jason Dong, Ian Ziyar, Alexander Shi, Runze Zhang, Lianghong Zheng, Rui Hou, William Shi, Xin Fu, Yaou Duan, Viet A N Huu, Cindy Wen, Edward D Zhang, Charlotte L Zhang, Oulan Li, Xiaobo Wang, Michael A Singer, Xiaodong Sun, Jie Xu, Ali Tafreshi, M Anthony Lewis, Huimin Xia, and Kang Zhang. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell, 2018. ISSN 0092-8674. doi: 10.1016/j.cell.2018.02.010. URL https://doi.org/10.1016/j.cell.2018.02.010.
- Khan et al. [2009] Ali Nawaz Khan, Hamdan Al-Jahdali, Sarah Al-Ghanem, and Alaa Gouda. Reading chest radiographs in the critically ill (Part I): Normal chest radiographic appearance, instrumentation and complications from instrumentation. Annals of Thoracic Medicine, 4(2):75–87, 7 2009. ISSN 18171737. doi: 10.4103/1817-1737.49416.
- Kim [2020] Hyungjin Kim. Outbreak of novel coronavirus (COVID-19): What is the role of radiologists?, 2020. ISSN 14321084.
- Kim et al. [2017] Taeksoo Kim, Moonsu Cha, Hyunsoo Kim, Jung Kwon Lee, and Jiwon Kim. Learning to Discover Cross-Domain Relations with Generative Adversarial Networks, 3 2017. URL http://arxiv.org/abs/1703.05192.
- Kingma and Ba [2014] Diederik Kingma and Jimmy Ba. Adam: A Method for Stochastic Optimization. International Conference on Learning Representations, 2014. ISSN 09252312. doi: 10.1145/1830483.1830503. URL http://arxiv.org/abs/1412.6980.
- Kong and Agarwal [2020] Weifang Kong and Prachi P. Agarwal. Chest imaging appearance of COVID-19 infection. Radiology: Cardiothoracic Imaging, 2(1):e200028, January 2020. doi: 10.1148/ryct.2020200028. URL https://doi.org/10.1148/ryct.2020200028.
- Kostkova et al. [2016] Patty Kostkova, Helen Brewer, Simon de Lusignan, Edward Fottrell, Ben Goldacre, Graham Hart, Phil Koczan, Peter Knight, Corinne Marsolier, Rachel A. McKendry, Emma Ross, Angela Sasse, Ralph Sullivan, Sarah Chaytor, Olivia Stevenson, Raquel Velho, and John Tooke. Who Owns the Data? Open Data for Healthcare. Frontiers in Public Health, 4:7, 2 2016. ISSN 2296-2565. doi: 10.3389/fpubh.2016.00007. URL http://journal.frontiersin.org/Article/10.3389/fpubh.2016.00007/abstract.
- Lakhani and Sundaram [2017] Paras Lakhani and Baskaran Sundaram. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology, 284(2):574–582, 8 2017. ISSN 0033-8419. doi: 10.1148/radiol.2017162326. URL http://pubs.rsna.org/doi/10.1148/radiol.2017162326.
- Larochelle et al. [2008] Hugo Larochelle, Dumitru Erhan, and Yoshua Bengio. Zero-data Learning of New Tasks. In Association for the Advancement of Artificial Intelligence, 2008.
- Lee and Yoon [2017] Choong Ho Lee and Hyung Jin Yoon. Medical big data: Promise and challenges. Kidney Research and Clinical Practice, 36(1):3–11, 3 2017. ISSN 22119140. doi: 10.23876/j.krcp.2017.36.1.3.
- Lee et al. [2020] Nan-Yao Lee, Chia-Wen Li, Huey-Pin Tsai, Po-Lin Chen, Ling-Shan Syue, Ming-Chi Li, Chin-Shiang Tsai, Ching-Lung Lo, Po-Ren Hsueh, and Wen-Chien Ko. A case of COVID-19 and pneumonia returning from macau in taiwan: Clinical course and anti-SARS-CoV-2 IgG dynamic. Journal of Microbiology, Immunology and Infection, March 2020. doi: 10.1016/j.jmii.2020.03.003. URL https://doi.org/10.1016/j.jmii.2020.03.003.
- Li et al. [2020] Matthew D. Li, Nishaeminth Thumbavanam Arun, Mishka Gidwani, Ken Chang, Francis Deng, Brent P. Little, Dexter P. Mendoza, Min Lang, Susanna I. Lee, Aileen O’Shea, Anushri Parakh, Praveer Singh, and Jayashree Kalpathy-Cramer. Automated Assessment and Tracking of COVID-19 Pulmonary Disease Severity on Chest Radiographs using Convolutional Siamese Neural Networks. Radiology: Artificial Intelligence, 2(4):e200079, 7 2020. ISSN 2638-6100. doi: 10.1148/ryai.2020200079. URL http://pubs.rsna.org/doi/10.1148/ryai.2020200079.
- Lim et al. [2020] Jaegyun Lim, Seunghyun Jeon, Hyun-Young Shin, Moon Jung Kim, Yu Min Seong, Wang Jun Lee, Kang-Won Choe, Yu Min Kang, Baeckseung Lee, and Sang-Joon Park. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. Journal of Korean Medical Science, 35(6), 2020. doi: 10.3346/jkms.2020.35.e79. URL https://doi.org/10.3346/jkms.2020.35.e79.
- Liu et al. [2020a] Ying-Chu Liu, Ching-Hui Liao, Chin-Fu Chang, Chu-Chung Chou, and Yan-Ren Lin. A locally transmitted case of SARS-CoV-2 infection in taiwan. New England Journal of Medicine, 382(11):1070–1072, March 2020a. doi: 10.1056/nejmc2001573. URL https://doi.org/10.1056/nejmc2001573.
- Liu et al. [2020b] YUjian Liu, Jianquan zhong, Hao Feng, and Minli Lv. Resolving COVID-19 pneumonia over time, 2020b. URL https://doi.org/10.1148/cases.20201394.
- Lyman [2020] Eric Lyman. Italian doctors turn to Chinese A.I. to speed up detection, 4 2020. URL https://fortune.com/2020/04/04/italy-coronavirus-symptoms-test-ai-china-covid-19/.
- Maguolo and Nanni [2020] Gianluca Maguolo and Loris Nanni. A Critic Evaluation of Methods for COVID-19 Automatic Detection from X-Ray Images. 4 2020. URL http://arxiv.org/abs/2004.12823.
- Majkowska et al. [2019] Anna Majkowska, Sid Mittal, David F. Steiner, Joshua J. Reicher, Scott Mayer McKinney, Gavin E. Duggan, Krish Eswaran, Po-Hsuan Cameron Chen, Yun Liu, Sreenivasa Raju Kalidindi, Alexander Ding, Greg S. Corrado, Daniel Tse, and Shravya Shetty. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-adjudicated Reference Standards and Population-adjusted Evaluation. Radiology, page 191293, 12 2019. ISSN 0033-8419. doi: 10.1148/radiol.2019191293. URL http://pubs.rsna.org/doi/10.1148/radiol.2019191293.
- Mastaglio et al. [2020] Sara Mastaglio, Annalisa Ruggeri, Antonio M. Risitano, Piera Angelillo, Despina Yancopoulou, Dimitrios C. Mastellos, Markus Huber-Lang, Simona Piemontese, Andrea Assanelli, Cecilia Garlanda, John D. Lambris, and Fabio Ciceri. The first case of covid-19 treated with the complement c3 inhibitor amy-101. Clinical Immunology, 2020. doi: 10.1016/j.clim.2020.108450.
- McCall [2020] Becky McCall. COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread. The Lancet Digital Health, 2020. ISSN 25897500. doi: 10.1016/s2589-7500(20)30054-6.
- McInnes et al. [2018] Leland McInnes, John Healy, and James Melville. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv preprint arXiv:1802.03426, 2 2018. URL http://arxiv.org/abs/1802.03426.
- Millán-Oñate et al. [2020] José Millán-Oñate, William Millan, Luis Alfonso Mendoza, Carlos Guillermo Sánchez, Hugo Fernandez-Suarez, D. Katterine Bonilla-Aldana, and Alfonso J. Rodríguez-Morales. Successful recovery of COVID-19 pneumonia in a patient from colombia after receiving chloroquine and clarithromycin. Annals of Clinical Microbiology and Antimicrobials, 19(1), April 2020. doi: 10.1186/s12941-020-00358-y. URL https://doi.org/10.1186/s12941-020-00358-y.
- Minaee et al. [2020] Shervin Minaee, Rahele Kafieh, Milan Sonka, Shakib Yazdani, and Ghazaleh Jamalipour Soufi. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Medical Image Analysis, 65, 10 2020. ISSN 13618423. doi: 10.1016/j.media.2020.101794. URL http://arxiv.org/abs/2004.09363.
- Monfardini et al. [2020] Lorenzo Monfardini, Claudio Sallemi, Nicolò Gennaro, Vittorio Pedicini, and Claudio Bnà. Contribution of interventional radiology to the management of covid-19 patient. CardioVascular and Interventional Radiology, 2020. doi: 10.1007/s00270-020-02470-0.
- Motiian et al. [2017] Saeid Motiian, Quinn Jones, Mehdi Iranmanesh, and Gianfranco Doretto. Few-Shot Adversarial Domain Adaptation. In Neural Information Processing Systems, 2017.
- Mukherjee et al. [2020] Aveek Mukherjee, Mudassar Ahmad, and Douglas Frenia. A coronavirus disease 2019 (covid-19) patient with multifocal pneumonia treated with hydroxychloroquine. Cureus, 2020. doi: 10.7759/cureus.7473.
- Murphy et al. [2020] Keelin Murphy, Henk Smits, Arnoud J.G. Knoops, Michael B.J.M. Korst, Tijs Samson, Ernst T. Scholten, Steven Schalekamp, Cornelia M. Schaefer-Prokop, Rick H.H.M. Philipsen, Annet Meijers, Jaime Melendez, Bram van Ginneken, and Matthieu Rutten. COVID-19 on Chest Radiographs: A Multireader Evaluation of an Artificial Intelligence System. Radiology, 296(3):E166–E172, 9 2020. ISSN 15271315. doi: 10.1148/radiol.2020201874. URL /pmc/articles/PMC7437494/?report=abstracthttps://www.ncbi.nlm.nih.gov/pmc/articles/PMC7437494/.
- Nair et al. [2020] A. Nair, J. C.L. Rodrigues, S. Hare, A. Edey, A. Devaraj, J. Jacob, A. Johnstone, R. McStay, Erika Denton, and G. Robinson. A British Society of Thoracic Imaging statement: considerations in designing local imaging diagnostic algorithms for the COVID-19 pandemic, 5 2020. ISSN 1365229X.
- Nakamura et al. [2020] Kazuha Nakamura, Mayu Hikone, Hiroshi Shimizu, Yusuke Kuwahara, Maki Tanabe, Mioko Kobayashi, Takuto Ishida, Kazuhiro Sugiyama, Takuya Washino, Naoya Sakamoto, and Yuichi Hamabe. A sporadic covid-19 pneumonia treated with extracorporeal membrane oxygenation in tokyo, japan: A case report. Journal of Infection and Chemotherapy, 2020. doi: 10.1016/j.jiac.2020.03.018.
- Ng et al. [2020] Ming-Yen Ng, Elaine YP Lee, Jin Yang, Fangfang Yang, Xia Li, Hongxia Wang, Macy Mei-sze Lui, Christine Shing-Yen Lo, Barry Leung, Pek-Lan Khong, Christopher Kim-Ming Hui, Kwok-yung Yuen, and Michael David Kuo. Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiology: Cardiothoracic Imaging, 2 2020. ISSN 2638-6135. doi: 10.1148/ryct.2020200034. URL http://pubs.rsna.org/doi/10.1148/ryct.2020200034.
- Ong et al. [2020] Eugenia Ziying Ong, Yvonne Fu Zi Chan, Wan Ying Leong, Natalie Mei Ying Lee, Shirin Kalimuddin, Salahudeen Mohamed Haja Mohideen, Kian Sing Chan, Anthony Tanoto Tan, Antonio Bertoletti, Eng Eong Ooi, and Jenny Guek Hong Low. A dynamic immune response shapes covid-19 progression. Cell Host & Microbe, 2020. doi: 10.1016/j.chom.2020.03.021.
- Paul et al. [2004] Narinder S. Paul, Heidi Roberts, Jagdish Butany, TaeBong Chung, Wayne Gold, Sangeeta Mehta, Eli Konen, Anuradha Rao, Yves Provost, Harry H. Hong, Leon Zelovitsky, and Gordon L. Weisbrod. Radiologic pattern of disease in patients with severe acute respiratory syndrome: The toronto experience. RadioGraphics, 24(2):553–563, March 2004. doi: 10.1148/rg.242035193. URL https://doi.org/10.1148/rg.242035193.
- Pedregosa et al. [2011] F Pedregosa, G Varoquaux, A Gramfort, V Michel, B Thirion, O Grisel, M Blondel, P Prettenhofer, R Weiss, V Dubourg, J Vanderplas, A Passos, D Cournapeau, M Brucher, M Perrot, and E Duchesnay. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12:2825–2830, 2011.
- Peng et al. [2020] Y Peng, YX Tang, S Lee, Zhu Y, RM Summers, and Z Lu. COVID-19-CT-CXR: a freely accessible and weakly labeled chest X-ray and CT image collection on COVID-19 from the biomedical literature. https://github.com/ncbi-nlp/COVID-19-CT-CXR., 2020.
- Pereira et al. [2020] Rodolfo M. Pereira, Diego Bertolini, Lucas O. Teixeira, Carlos N. Silla, and Yandre M.G. Costa. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Computer Methods and Programs in Biomedicine, 194:105532, 10 2020. ISSN 18727565. doi: 10.1016/j.cmpb.2020.105532.
- Phan et al. [2020] Lan T. Phan, Thuong V. Nguyen, Quang C. Luong, Thinh V. Nguyen, Hieu T. Nguyen, Hung Q. Le, Thuc T. Nguyen, Thang M. Cao, and Quang D. Pham. Importation and human-to-human transmission of a novel coronavirus in vietnam. New England Journal of Medicine, 382(9):872–874, February 2020. doi: 10.1056/nejmc2001272. URL https://doi.org/10.1056/nejmc2001272.
- Poggiali et al. [2020] Erika Poggiali, Alessandro Dacrema, Davide Bastoni, Valentina Tinelli, Elena Demichele, Pau Mateo Ramos, Teodoro Marcianò, Matteo Silva, Andrea Vercelli, and Andrea Magnacavallo. Can Lung US Help Critical Care Clinicians in the Early Diagnosis of Novel Coronavirus (COVID-19) Pneumonia?, 6 2020. ISSN 15271315.
- Prince and Sergel [2020] Garrett Prince and Michelle Sergel. Persistent hiccups as an atypical presenting complaint of covid-19. The American Journal of Emergency Medicine, 2020. doi: 10.1016/j.ajem.2020.04.045.
- Putha et al. [2018b] Preetham Putha, Manoj Tadepalli, Bhargava Reddy, Tarun Raj, Justy Antony Chiramal, Shalini Govil, Namita Sinha, Manjunath KS, Sundeep Reddivari, Ammar Jagirdar, Pooja Rao, and Prashant Warier. Can Artificial Intelligence Reliably Report Chest X-Rays?: Radiologist Validation of an Algorithm trained on 2.3 Million X-Rays. arxiv, 7 2018b. URL http://arxiv.org/abs/1807.07455.
- Putha et al. [2018a] Preetham Putha, Manoj Tadepalli, Bhargava Reddy, Tarun Raj, Justy Antony Chiramal, Shalini Govil, Namita Sinha, Manjunath KS, Sundeep Reddivari, Ammar Jagirdar, Pooja Rao, and Prashant Warier. Can artificial intelligence reliably report chest x-rays?: Radiologist validation of an algorithm trained on 2.3 million x-rays, 2018a.
- Qian et al. [2020] Lijuan Qian, Jie Yu, and Heshui Shi. Severe Acute Respiratory Disease in a Huanan Seafood Market Worker: Images of an Early Casualty. Radiology: Cardiothoracic Imaging, 2(1):e200033, 2 2020. ISSN 2638-6135. doi: 10.1148/ryct.2020200033. URL http://pubs.rsna.org/doi/10.1148/ryct.2020200033.
- Radbel et al. [2020] Jared Radbel, Navaneeth Narayanan, and Pinki J. Bhatt. Use of tocilizumab for covid-19-induced cytokine release syndrome. Chest, 2020. doi: 10.1016/j.chest.2020.04.024.
- Raj [2020] Tarun Raj. Re-purposing qXR for COVID-19, 2020. URL http://blog.qure.ai/notes/chest-xray-AI-qxr-for-covid-19.
- Rajpurkar et al. [2017] Pranav Rajpurkar, Jeremy Irvin, Kaylie Zhu, Brandon Yang, Hershel Mehta, Tony Duan, Daisy Ding, Aarti Bagul, Curtis Langlotz, Katie Shpanskaya, Matthew P. Lungren, and Andrew Y. Ng. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv:1711.05225 [cs, stat], November 2017. URL http://arxiv.org/abs/1711.05225. arXiv: 1711.05225.
- Rajpurkar et al. [2018] Pranav Rajpurkar, Jeremy Irvin, Robyn L. Ball, Kaylie Zhu, Brandon Yang, Hershel Mehta, Tony Duan, Daisy Ding, Aarti Bagul, Curtis P. Langlotz, Bhavik N. Patel, Kristen W. Yeom, Katie Shpanskaya, Francis G. Blankenberg, Jayne Seekins, Timothy J. Amrhein, David A. Mong, Safwan S. Halabi, Evan J. Zucker, Andrew Y. Ng, and Matthew P. Lungren. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLOS Medicine, 15(11):e1002686, 11 2018. ISSN 1549-1676. doi: 10.1371/journal.pmed.1002686. URL http://dx.plos.org/10.1371/journal.pmed.1002686.
- Ren et al. [2019] Mengye Ren, Renjie Liao, Ethan Fetaya, and Richard S Zemel. Incremental Few-Shot Learning with Attention Attractor Networks. In Neural Information Processing Systems, pages 5275–5285, 2019.
- Rodriguez et al. [2020] Jose A. Rodriguez, Heysu Rubio-Gomez, Alejandra A. Roa, N. Miller, and Paula A. Eckardt. Co-infection with SARS-COV-2 and parainfluenza in a young adult patient with pneumonia: Case report. IDCases, 20:e00762, 2020. doi: 10.1016/j.idcr.2020.e00762. URL https://doi.org/10.1016/j.idcr.2020.e00762.
- Ross et al. [2017] Andrew Ross, Michael C Hughes, and Finale Doshi-Velez. Right for the Right Reasons: Training Differentiable Models by Constraining their Explanations. In International Joint Conference on Artificial Intelligence, 2017. URL https://github.com/dtak/rrr.
- Roth et al. [2020] Karsten Roth, Timo Milbich, Samarth Sinha, Prateek Gupta, Björn Ommer, and Joseph Paul Cohen. Revisiting Training Strategies and Generalization Performance in Deep Metric Learning. In International Conference of Machine Learning, 2 2020. URL http://arxiv.org/abs/2002.08473.
- Rubin et al. [2020] Geoffrey D. Rubin, Linda B. Haramati, Jeffrey P. Kanne, Neil W. Schluger, Jae-Joon Yim, Deverick J. Anderson, Talissa Altes, Sujal R. Desai, Jin Mo Goo, Yoshikazu Inoue, Fengming Luo, Mathias Prokop, Luca Richeldi, Noriyuki Tomiyama, Ann N. Leung, Christopher J. Ryerson, Nicola Sverzellati, Suhail Raoof, Annalisa Volpi, Ian B. K. Martin, Christina Kong, Andrew Bush, Jonathan Goldin, Marc Humbert, Hans-Ulrich Kauczor, Peter J. Mazzone, Martine Remy-Jardin, Cornelia M. Schaefer-Prokop, and Athol U. Wells. The Role of Chest Imaging in Patient Management during the COVID-19 Pandemic: A Multinational Consensus Statement from the Fleischner Society. Radiology, 2020. ISSN 0033-8419. doi: 10.1148/radiol.2020201365. URL http://pubs.rsna.org/doi/10.1148/radiol.2020201365.
- Rubin et al. [2018] Jonathan Rubin, Deepan Sanghavi, Claire Zhao, Kathy Lee, Ashequl Qadir, and Minnan Xu-Wilson. Large Scale Automated Reading of Frontal and Lateral Chest X-Rays using Dual Convolutional Neural Networks. In Conference on Machine Intelligence in Medical Imaging, 4 2018. URL http://arxiv.org/abs/1804.07839.
- Salehi et al. [2020] Sana Salehi, Aidin Abedi, Sudheer Balakrishnan, and Ali Gholamrezanezhad. Coronavirus Disease 2019 (COVID-19): A Systematic Review of Imaging Findings in 919 Patients. American Journal of Roentgenology, pages 1–7, 3 2020. ISSN 0361-803X. doi: 10.2214/AJR.20.23034. URL https://www.ajronline.org/doi/10.2214/AJR.20.23034.
- Sánchez-Oro et al. [2020] Raquel Sánchez-Oro, Julio Torres Nuez, and Gloria Martínez-Sanz. Radiological findings for diagnosis of sars-cov-2 pneumonia (covid-19). Medicina Clínica (English Edition), 2020. doi: 10.1016/j.medcle.2020.03.004.
- Selvan et al. [2020] Raghavendra Selvan, Erik B. Dam, Sofus Rischel, Kaining Sheng, Mads Nielsen, and Akshay Pai. Lung Segmentation from Chest X-rays using Variational Data Imputation. 5 2020. URL http://arxiv.org/abs/2005.10052.
- Seyyed-Kalantari et al. [2020] Laleh Seyyed-Kalantari, Guanxiong Liu, Matthew McDermott, and Marzyeh Ghassemi. CheXclusion: Fairness gaps in deep chest X-ray classifiers. 2 2020. URL http://arxiv.org/abs/2003.00827.
- Shafaei et al. [2018] Alireza Shafaei, Mark Schmidt, and James J. Little. Does Your Model Know the Digit 6 Is Not a Cat? A Less Biased Evaluation of "Outlier" Detectors. arxiv, 9 2018. URL http://arxiv.org/abs/1809.04729.
- Shamout et al. [2020] Farah E. Shamout, Yiqiu Shen, Nan Wu, Aakash Kaku, Jungkyu Park, Taro Makino, Stanisław Jastrzȩbski, Duo Wang, Ben Zhang, Siddhant Dogra, Meng Cao, Narges Razavian, David Kudlowitz, Lea Azour, William Moore, Yvonne W. Lui, Yindalon Aphinyanaphongs, Carlos Fernandez-Granda, and Krzysztof J. Geras. An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department, 8 2020. ISSN 2331-8422. URL https://github.com/nyukat/COVID-19_prognosis.
- Shi et al. [2020] Heshui Shi, Xiaoyu Han, and Chuansheng Zheng. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in wuhan, china. Radiology, 295(1):20–20, April 2020. doi: 10.1148/radiol.2020200269. URL https://doi.org/10.1148/radiol.2020200269.
- Siddamreddy et al. [2020] Suman Siddamreddy, Ramakrishna Thotakura, Vasuki Dandu, Sruthi Kanuru, and Sreenath Meegada. Corona virus disease 2019 (covid-19) presenting as acute st elevation myocardial infarction. Cureus, 2020. doi: 10.7759/cureus.7782.
- Signoroni et al. [2020] Alberto Signoroni, Mattia Savardi, Sergio Benini, Nicola Adami, Riccardo Leonardi, Paolo Gibellini, Filippo Vaccher, Marco Ravanelli, Andrea Borghesi, Roberto Maroldi, and Davide Farina. End-to-end learning for semiquantitative rating of COVID-19 severity on Chest X-rays. arXiv:2006.04603, 6 2020. URL http://arxiv.org/abs/2006.04603.
- Silverstein et al. [2020] William Kyle Silverstein, Lynfa Stroud, Graham Edward Cleghorn, and Jerome Allen Leis. First imported case of 2019 novel coronavirus in canada, presenting as mild pneumonia. The Lancet, 395(10225):734, February 2020. doi: 10.1016/s0140-6736(20)30370-6. URL https://doi.org/10.1016/s0140-6736(20)30370-6.
- Simonite [2020] Tom Simonite. Chinese Hospitals Deploy AI to Help Diagnose COVID-19, 2 2020. URL https://www.wired.com/story/chinese-hospitals-deploy-ai-help-diagnose-covid-19/.
- SINGH et al. [2020] RAJAT SINGH, CHRISTOPHER DOMENICO, SRIRAM D. RAO, KIMBERLY URGO, STUART B. PRENNER, JOYCE W. WALD, PAVAN ATLURI, and EDO Y. BIRATI. Novel coronavirus disease 2019 in a patient on durable left ventricular assist device support. Journal of Cardiac Failure, 2020. doi: 10.1016/j.cardfail.2020.04.007.
- Singla et al. [2020] Sumedha Singla, Brian Pollack, Junxiang Chen, and Kayhan Batmanghelich. Explanation by Progressive Exaggeration. In International Conference on Learning Representations, 11 2020. URL http://arxiv.org/abs/1911.00483.
- Sivakorn et al. [2020] Chaisith Sivakorn, Viravarn Luvira, Sant Muangnoicharoen, Pittaya Piroonamornpun, Tharawit Ouppapong, Anek Mungaomklang, and Sopon Iamsirithaworn. Case report: Walking pneumonia in novel coronavirus disease (COVID-19): Mild symptoms with marked abnormalities on chest imaging. The American Journal of Tropical Medicine and Hygiene, 102(5):940–942, May 2020. doi: 10.4269/ajtmh.20-0203. URL https://doi.org/10.4269/ajtmh.20-0203.
- Snell et al. [2017] Jake Snell, Kevin Swersky, and Twitter Richard Zemel. Prototypical Networks for Few-shot Learning. In Neural Information Processing Systems, 2017.
- Song et al. [2020a] Fengxiang Song, Nannan Shi, Fei Shan, Zhiyong Zhang, Jie Shen, Hongzhou Lu, Yun Ling, Yebin Jiang, and Yuxin Shi. Emerging 2019 novel coronavirus (2019-NCoV) pneumonia. Radiology, 295(1):210–217, 2 2020a. ISSN 15271315. doi: 10.1148/radiol.2020200274.
- Song et al. [2020b] Jehun Song, Seongmin Kang, Seung Won Choi, Kwang Won Seo, Sunggun Lee, Min Wook So, and Doo-Ho Lim. Coronavirus disease 19 (covid-19) complicated with pneumonia in a patient with rheumatoid arthritis receiving conventional disease-modifying antirheumatic drugs. Rheumatology International, 2020b. doi: 10.1007/s00296-020-04584-7.
- Stirenko et al. [2018] S. Stirenko, Y. Kochura, O. Alienin, O. Rokovyi, Y. Gordienko, P. Gang, and W. Zeng. Chest x-ray analysis of tuberculosis by deep learning with segmentation and augmentation. In 2018 IEEE 38th International Conference on Electronics and Nanotechnology (ELNANO), pages 422–428, 2018.
- Strickland [2020] Eliza Strickland. AI Can Help Hospitals Triage COVID-19 Patients, 2020. URL https://spectrum.ieee.org/the-human-os/artificial-intelligence/medical-ai/ai-can-help-hospitals-triage-covid19-patients.
- Taghanaki et al. [2019] Saeid Asgari Taghanaki, Mohammad Havaei, Tess Berthier, Francis Dutil, Lisa Di Jorio, Ghassan Hamarneh, and Yoshua Bengio. InfoMask: Masked Variational Latent Representation to Localize Chest Disease. In Medical Image Computing and Computer Assisted Interventions. Springer, 3 2019. ISBN 9783030322250. doi: 10.1007/978-3-030-32226-7{\_}82. URL http://arxiv.org/abs/1903.11741.
- Tartaglione et al. [2020] Enzo Tartaglione, Carlo Alberto Barbano, Claudio Berzovini, Marco Calandri, and Marco Grangetto. Unveiling COVID-19 from Chest X-ray with deep learning: a hurdles race with small data. 4 2020. URL http://arxiv.org/abs/2004.05405.
- Tay and Harwood [2020] Hui Sian Tay and Rowan Harwood. Atypical presentation of covid-19 in a frail older person. Age and Ageing, 2020. doi: 10.1093/ageing/afaa068.
- Thevarajan et al. [2020] Irani Thevarajan, Thi H. O. Nguyen, Marios Koutsakos, Julian Druce, Leon Caly, Carolien E. van de Sandt, Xiaoxiao Jia, Suellen Nicholson, Mike Catton, Benjamin Cowie, Steven Y. C. Tong, Sharon R. Lewin, and Katherine Kedzierska. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nature Medicine, March 2020. doi: 10.1038/s41591-020-0819-2. URL https://doi.org/10.1038/s41591-020-0819-2.
- Thille et al. [2013] Arnaud W. Thille, Jean Christophe M. Richard, and Laurent Brochard. The decision to extubate in the intensive care unit, 6 2013. ISSN 1073449X. URL http://www.atsjournals.org/doi/abs/10.1164/rccm.201208-1523CI.
- Tian et al. [2020a] Sufang Tian, Yong Xiong, Huan Liu, Li Niu, Jianchun Guo, Meiyan Liao, and Shu-Yuan Xiao. Pathological study of the 2019 novel coronavirus disease (covid-19) through postmortem core biopsies. Modern Pathology, 2020a. doi: 10.1038/s41379-020-0536-x.
- Tian et al. [2020b] Yonglong Tian, Yue Wang, Dilip Krishnan, Joshua B Tenenbaum, and Phillip Isola. Rethinking Few-Shot Image Classification: a Good Embedding Is All You Need? 2020b. URL http://github.com/WangYueFt/http://arxiv.org/abs/2003.11539.
- Tobin and Manthous [2017] Martin Tobin and Constantine Manthous. Mechanical Ventilation. Technical report, American Thoracic Society, 2017. URL http://www.caregiver.org.
- Tonelli et al. [2020] R. Tonelli, A. Iattoni, M. Girardis, L. De Pietri, E. Clini, and C. Mussini. Never give up: Lesson learned from a severe covid-19 patient. Pulmonology, 2020. doi: 10.1016/j.pulmoe.2020.04.012.
- Varga et al. [2020] Zsuzsanna Varga, Andreas J Flammer, Peter Steiger, Martina Haberecker, Rea Andermatt, Annelies S Zinkernagel, Mandeep R Mehra, Reto A Schuepbach, Frank Ruschitzka, and Holger Moch. Endothelial cell infection and endotheliitis in covid-19. The Lancet, 395(10234):1417–1418, 2020.
- Viviano et al. [2019] Joseph D. Viviano, Becks Simpson, Francis Dutil, Yoshua Bengio, and Joseph Paul Cohen. Saliency is a Possible Red Herring When Diagnosing Poor Generalization. arxiv:1910.00199, 10 2019. URL http://arxiv.org/abs/1910.00199.
- Vollono et al. [2020] Catello Vollono, Eleonora Rollo, Marina Romozzi, Giovanni Frisullo, Serenella Servidei, Alberto Borghetti, and Paolo Calabresi. Focal status epilepticus as unique clinical feature of covid-19: A case report. Seizure, 2020. doi: 10.1016/j.seizure.2020.04.009.
- Vu and Vu [2020] Dan Vu and Peter Vu. COVID-19 pneumonia, 2020. URL https://doi.org/10.1148/cases.20201815.
- Wadman et al. [2020] M Wadman, J Couzin-Frankel, J Kaiser, and C Matacic. How does coronavirus kill. Clinicians trace a ferocious rampage through the body, from brain to toes, pages 1502–1503, 2020.
- WANG et al. [2020] Lili WANG, Junfeng Li, and Junqiang Lei. COVID-19 pneumonia-disease progression over time, 2020. URL https://doi.org/10.1148/cases.20201559.
- Wang and Wong [2020] Linda Wang and Alexander Wong. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. 3 2020. URL http://arxiv.org/abs/2003.09871.
- Wang et al. [2017] Xiaosong Wang, Yifan Peng, Le Lu, Zhiyong Lu, Mohammadhadi Bagheri, and Ronald M. Summers. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Computer Vision and Pattern Recognition, 2017. doi: 10.1109/CVPR.2017.369. URL http://arxiv.org/abs/1705.02315.
- Wang et al. [2019] Yaqing Wang, Quanming Yao, James Kwok, and Lionel M. Ni. Generalizing from a Few Examples: A Survey on Few-Shot Learning. ACM Computing Surveys, 4 2019. URL http://arxiv.org/abs/1904.05046.
- Wanshu Zhang and Heshui Shi [2020] MD Wanshu Zhang and MD Heshui Shi. Evolving COVID-19 pneumonia, 2020. URL https://doi.org/10.1148/cases.20201558.
- Wei et al. [2020] Jiangping Wei, Huaxiang Xu, Jingliang Xiong, Qinglin Shen, Bing Fan, Chenglong Ye, Wentao Dong, and Fangfang Hu. 2019 novel coronavirus (COVID-19) pneumonia: Serial computed tomography findings. Korean Journal of Radiology, 21(4):501, 2020. doi: 10.3348/kjr.2020.0112. URL https://doi.org/10.3348/kjr.2020.0112.
- Winther et al. [2020a] Hinrich B. Winther, Hans Laser, Svetlana Gerbel, Sabine K. Maschke, Jan B. Hinrichs, Jens Vogel-Claussen, Frank K. Wacker, Marius M. Höper, and Bernhard C. Meyer. Covid-19 image repository, 2020a. URL https://figshare.com/articles/COVID-19_Image_Repository/12275009.
- Winther et al. [2020b] Hinrich B. Winther, Hans Laser, Svetlana Gerbel, Sabine K. Maschke, Jan B. Hinrichs, Jens Vogel-Claussen, Frank K. Wacker, Marius M. Höper, and Bernhard C. Meyer. COVID-19 Image Repository, 2020b.
- Wong et al. [2019] Ho Yuen Frank Wong, Hiu Yin Sonia Lam, Ambrose Ho Tung Fong, Siu Ting Leung, Thomas Wing Yan Chin, Christine Shing Yen Lo, Macy Mei Sze Lui, Jonan Chun Yin Lee, Keith Wan Hang Chiu, Tom Chung, Elaine Yuen Phin Lee, Eric Yuk Fai Wan, Fan Ngai Ivan Hung, Tina Poy Wing Lam, Michael Kuo, and Ming Yen Ng. Frequency and Distribution of Chest Radiographic Findings in COVID-19 Positive Patients. Radiology, 3 2019. ISSN 15271315. doi: 10.1148/radiol.2020201160.
- Wong et al. [2003] K. T. Wong, Gregory E. Antonio, David S.C. Hui, Nelson Lee, Edmund H.Y. Yuen, Alan Wu, C. B. Leung, T. H. Rainer, Peter Cameron, Sydney S.C. Chung, Joseph J.Y. Sung, and Anil T. Ahuja. Severe acute respiratory syndrome: Radiographic appearances and pattern of progression in 138 patients. Radiology, 228(2):401–406, 8 2003. ISSN 00338419. doi: 10.1148/radiol.2282030593.
- Wong et al. [2020] S.C.Y. Wong, R.T-S. Kwong, T.C. Wu, J.W.M. Chan, M.Y. Chu, S.Y. Lee, H.Y. Wong, and D.C. Lung. Risk of nosocomial transmission of coronavirus disease 2019: an experience in a general ward setting in hong kong. Journal of Hospital Infection, 2020. doi: 10.1016/j.jhin.2020.03.036.
- Woznitza et al. [2020] N. Woznitza, A. Nair, and S.S. Hare. Covid-19: A case series to support radiographer preliminary clinical evaluation. Radiography, 2020. doi: 10.1016/j.radi.2020.04.002.
- Wu et al. [2020] Jian Wu, Jun Liu, Xinguo Zhao, Chengyuan Liu, Wei Wang, Dawei Wang, Wei Xu, Chunyu Zhang, Jiong Yu, Bin Jiang, Hongcui Cao, and Lanjuan Li. Clinical characteristics of imported cases of COVID-19 in jiangsu province: A multicenter descriptive study. Clinical Infectious Diseases, February 2020. doi: 10.1093/cid/ciaa199. URL https://doi.org/10.1093/cid/ciaa199.
- Wynants et al. [2020] Laure Wynants, Ben Van Calster, Marc M.J. Bonten, Gary S. Collins, Thomas P.A. Debray, Maarten De Vos, Maria C. Haller, Georg Heinze, Karel G.M. Moons, Richard D. Riley, Ewoud Schuit, Luc J.M. Smits, Kym I.E. Snell, Ewout W. Steyerberg, Christine Wallisch, and Maarten Van Smeden. Prediction models for diagnosis and prognosis of COVID-19 infection: Systematic review and critical appraisal. The BMJ, 369, 4 2020. ISSN 17561833. doi: 10.1136/bmj.m1328.
- Xu et al. [2020] Z Xu, L Shi, J Zhang, L Huang, C Zhang, H Liu Bsc, J Song, F-S Wang, Y Wang, Y Tai, J Zhao, Fu-Sheng Wang, Jingmin Zhao, Zhe Xu, Lei Shi, Yijin Wang, Jiyuan Zhang, Lei Huang, Chao Zhang, Shuhong Liu, Peng Zhao, Hongxia Liu, Li Zhu, Yanhong Tai, Changqing Bai, Tingting Gao, Jinwen Song, Peng Xia, and Jinghui Dong. Case Report Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet Respiratory, 8:420–422, 2020. doi: 10.1016/S2213-2600(20)30076-X. URL https://doi.org/10.1016/.
- Yan et al. [2020] Li Yan, Hai-Tao Zhang, Yang Xiao, Maolin Wang, Chuan Sun, Jing Liang, Shusheng Li, Mingyang Zhang, Yuqi Guo, Ying Xiao, Xiuchuan Tang, Haosen Cao, Xi Tan, Niannian Huang, Bo Jiao, Ailin Luo, Zhiguo Cao, Hui Xu, and Ye Yuan. Prediction of criticality in patients with severe Covid-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan. medRxiv, 3 2020. doi: 10.1101/2020.02.27.20028027. URL http://medrxiv.org/content/early/2020/03/03/2020.02.27.20028027.abstract.
- Yoon et al. [2020] Soon Ho Yoon, Kyung Hee Lee, Jin Yong Kim, Young Kyung Lee, Hongseok Ko, Ki Hwan Kim, Chang Min Park, and Yun-Hyeon Kim. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): Analysis of nine patients treated in korea. Korean Journal of Radiology, 21(4):494, 2020. doi: 10.3348/kjr.2020.0132. URL https://doi.org/10.3348/kjr.2020.0132.
- Yu et al. [2020] Qian Yu, Yuancheng Wang, Shan Huang, Songqiao Liu, Zhen Zhou, Shijun Zhang, Zhen Zhao, Yizhou Yu, Yi Yang, and Shenghong Ju. Multicenter cohort study demonstrates more consolidation in upper lungs on initial CT increases the risk of adverse clinical outcome in COVID-19 patients. Theranostics, 10(12):5641–5648, 2020. doi: 10.7150/thno.46465. URL https://doi.org/10.7150/thno.46465.
- Zanin et al. [2020] Luca Zanin, Giorgio Saraceno, Pier Paolo Panciani, Giulia Renisi, Liana Signorini, Karol Migliorati, and Marco Maria Fontanella. Sars-cov-2 can induce brain and spine demyelinating lesions. Acta Neurochirurgica, 2020. doi: 10.1007/s00701-020-04374-x.
- Zhang et al. [2020] Jianpeng Zhang, Yutong Xie, Yi Li, Chunhua Shen, and Yong Xia. COVID-19 Screening on Chest X-ray Images Using Deep Learning based Anomaly Detection. 3 2020. URL http://arxiv.org/abs/2003.12338.
- Zhu et al. [2017] Jun Yan Zhu, Taesung Park, Phillip Isola, and Alexei A. Efros. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In International Conference on Computer Vision, volume 2017-Octob, 2017. ISBN 9781538610329. doi: 10.1109/ICCV.2017.244. URL https://arxiv.org/pdf/1703.10593.pdf.
- Zhu et al. [2020] Na Zhu, Dingyu Zhang, Wenling Wang, Xingwang Li, Bo Yang, Jingdong Song, Xiang Zhao, Baoying Huang, Weifeng Shi, Roujian Lu, Peihua Niu, Faxian Zhan, Xuejun Ma, Dayan Wang, Wenbo Xu, Guizhen Wu, George F. Gao, and Wenjie Tan. A novel coronavirus from patients with pneumonia in china, 2019. New England Journal of Medicine, 2020. doi: 10.1056/nejmoa2001017.
- Zu et al. [2020] Zi Yue Zu, Meng Di Jiang, Peng Peng Xu, Wen Chen, Qian Qian Ni, Guang Ming Lu, and Long Jiang Zhang. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology, page 200490, 2 2020. ISSN 15271315. doi: 10.1148/radiol.2020200490.
Appendix
A Extra dataset statistics
Just considering PA, AP, and AP Supine views there are 367 JPEG files and 171 PNGs. All images are 8-bit except for 1 which is 16-bit.
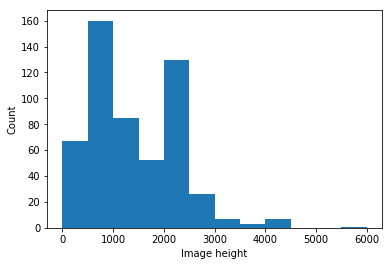
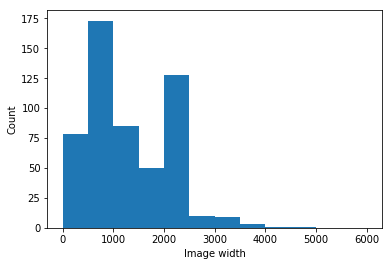
B Example images
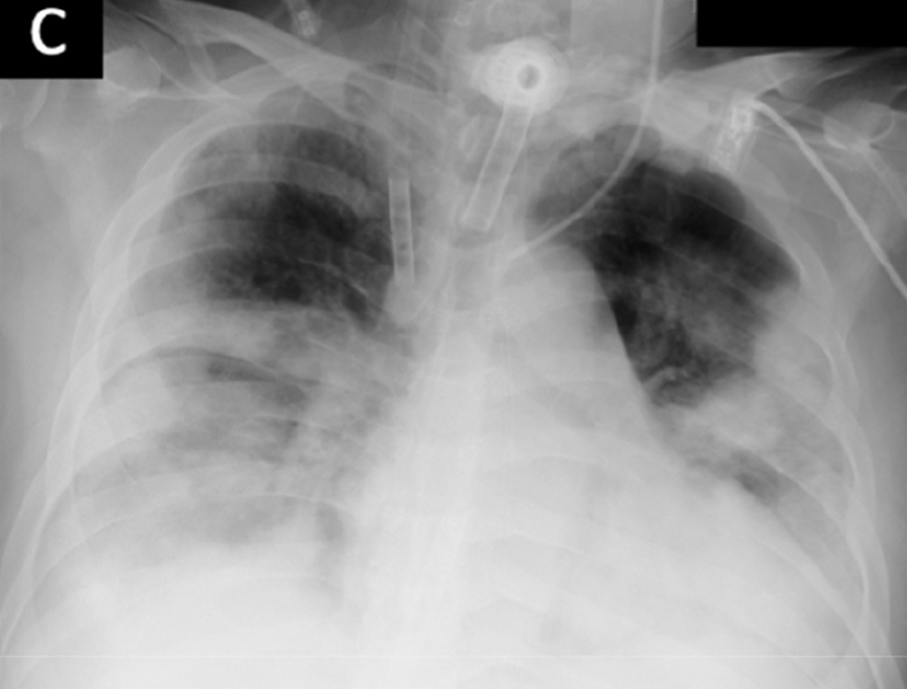
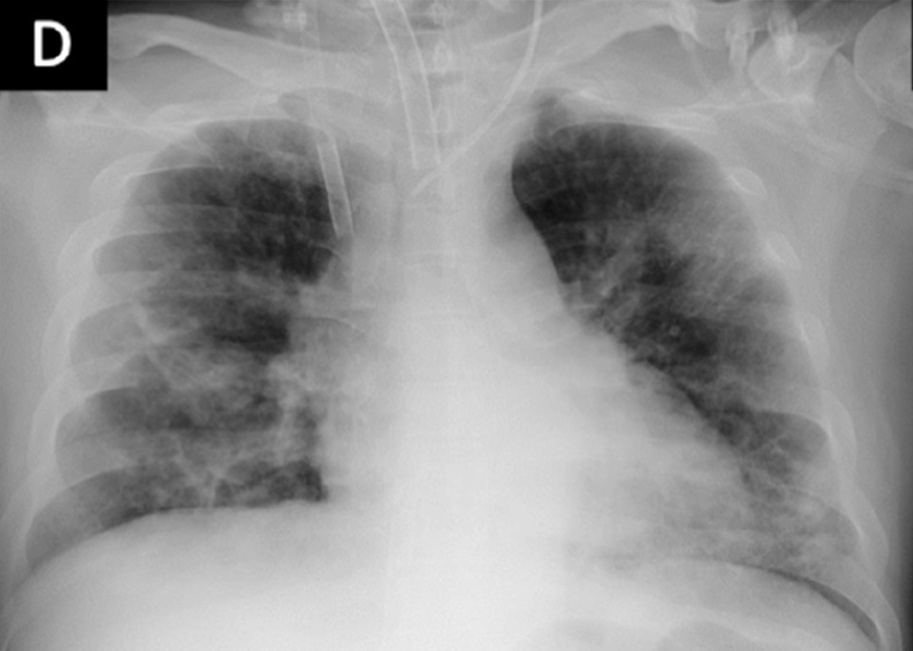
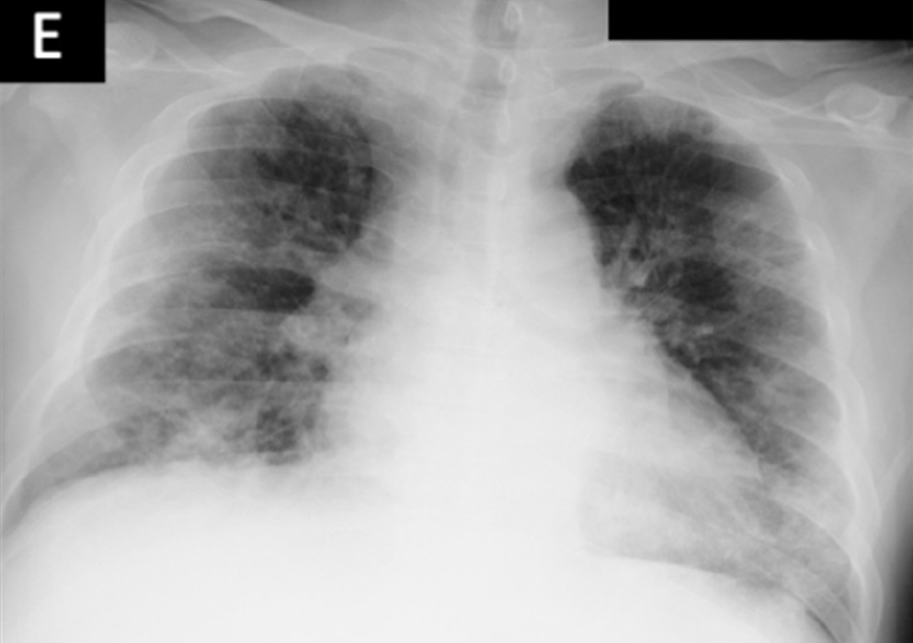

C Scraper details
Each scraper identifies relevant radiographs using the “search" feature of the site being scraped, and saves the images together with a csv file of the corresponding metadata. Internally, the scraper uses Selenium to visit each page of the search results, saving metadata and images from any new cases. Images are downloaded at the highest resolution available. Data from each case is converted to a single, interoperable JSON format. Finally, the needed metadata is exported as a csv file.
Currently, the scrapers only retrieve metadata from structured fields that can be easily detected on webpages, such as “Age", “Sex", “Imaging Notes", and “Clinical Notes." Other clinically important information (such as whether the patient went to the ICU or was intubated) is extracted by human annotators.
Care has been taken to use these websites’ resources conservatively and avoid creating a burden on their servers. Crawling is done in a lazy manner, so that no pages are requested until they can be used. Also, within each scraping session, each web page is kept open in an individual browser instance as long as it is needed to reduce re-requested resources .
C.1 Papers where images and clinical data are sourced
| Citation | PA/AP | AP Supine | L |
|---|---|---|---|
| Winther et al. [2020a] | 48 | 44 | 0 |
| Paul et al. [2004] | 11 | 0 | 0 |
| Wong et al. [2019] | 9 | 0 | 0 |
| Wong et al. [2003] | 5 | 0 | 0 |
| Cheng et al. [2020] | 4 | 0 | 0 |
| Hsih et al. [2020] | 4 | 0 | 0 |
| Woznitza et al. [2020] | 4 | 0 | 0 |
| Holshue et al. [2020] | 4 | 0 | 3 |
| Phan et al. [2020] | 4 | 0 | 0 |
| Ng et al. [2020] | 4 | 0 | 0 |
| Coimbra et al. [2020] | 3 | 0 | 0 |
| Song et al. [2020b] | 3 | 0 | 0 |
| Hiramatsu et al. [2020] | 3 | 0 | 0 |
| Ong et al. [2020] | 3 | 0 | 0 |
| Azekawa et al. [2020] | 3 | 0 | 0 |
| Sánchez-Oro et al. [2020] | 3 | 0 | 0 |
| Wu et al. [2020] | 3 | 0 | 0 |
| Qian et al. [2020] | 3 | 0 | 0 |
| Lim et al. [2020] | 3 | 0 | 0 |
| Yoon et al. [2020] | 3 | 0 | 0 |
| Sivakorn et al. [2020] | 3 | 0 | 0 |
| Borghesi and Maroldi [2020a] | 2 | 3 | 0 |
| jin Zhang et al. [2020] | 2 | 3 | 0 |
| Rodriguez et al. [2020] | 2 | 1 | 0 |
| Chen et al. [2020] | 2 | 0 | 0 |
| Cuong et al. [2020] | 2 | 0 | 0 |
| SINGH et al. [2020] | 2 | 0 | 0 |
| Hare et al. [2020] | 2 | 0 | 0 |
| Huang et al. [2020] | 2 | 0 | 0 |
| Lee et al. [2020] | 2 | 0 | 0 |
| Banerjee et al. [2020] | 2 | 0 | 0 |
| Vollono et al. [2020] | 2 | 0 | 0 |
| Tian et al. [2020a] | 2 | 0 | 0 |
| Thevarajan et al. [2020] | 2 | 0 | 0 |
| Liu et al. [2020a] | 2 | 0 | 0 |
| Cai et al. [2020] | 2 | 0 | 0 |
| Ahmed et al. [2020] | 2 | 0 | 0 |
| Fichera et al. [2020] | 1 | 7 | 0 |
| Ebrille et al. [2020] | 1 | 3 | 0 |
| Xu et al. [2020] | 1 | 2 | 0 |
| Dastan et al. [2020] | 1 | 2 | 0 |
| Monfardini et al. [2020] | 1 | 1 | 0 |
| Borghesi et al. [2020b] | 1 | 1 | 0 |
| Mastaglio et al. [2020] | 1 | 1 | 0 |
| Silverstein et al. [2020] | 1 | 0 | 0 |
| Aigner et al. [2020] | 1 | 0 | 0 |
| Wong et al. [2020] | 1 | 0 | 0 |
| Tay and Harwood [2020] | 1 | 0 | 0 |
| Liu et al. [2020b] | 1 | 0 | 0 |
| Wanshu Zhang and Heshui Shi [2020] | 1 | 0 | 0 |
| WANG et al. [2020] | 1 | 0 | 0 |
| Vu and Vu [2020] | 1 | 0 | 0 |
| Shi et al. [2020] | 1 | 0 | 0 |
| Song et al. [2020a] | 1 | 0 | 0 |
| Zu et al. [2020] | 1 | 0 | 0 |
| Kong and Agarwal [2020] | 1 | 0 | 0 |
| Millán-Oñate et al. [2020] | 1 | 0 | 0 |
| Jin et al. [2020] | 1 | 0 | 0 |
| Citation | PA/AP | AP Supine | L |
|---|---|---|---|
| Wei et al. [2020] | 1 | 0 | 0 |
| Allen et al. [2010] | 1 | 0 | 0 |
| An et al. [2020] | 1 | 0 | 0 |
| Yu et al. [2020] | 1 | 0 | 0 |
| Mukherjee et al. [2020] | 1 | 0 | 0 |
| Nakamura et al. [2020] | 0 | 5 | 0 |
| Zhu et al. [2020] | 0 | 2 | 0 |
| Bhatraju et al. [2020] | 0 | 2 | 0 |
| Siddamreddy et al. [2020] | 0 | 2 | 0 |
| Zanin et al. [2020] | 0 | 1 | 0 |
| Prince and Sergel [2020] | 0 | 1 | 0 |
| Fidan [2020] | 0 | 1 | 0 |
| Avula et al. [2020] | 0 | 1 | 0 |
| Radbel et al. [2020] | 0 | 1 | 0 |
| Tonelli et al. [2020] | 0 | 1 | 0 |
| Salehi et al. [2020] | 0 | 1 | 0 |
| Salehi et al. [2020] | 0 | 1 | 0 |
| Filatov et al. [2020] | 0 | 1 | 0 |
C.2 Output of models on each split
| # params | # test samples | Correlation | MAE | R^2 | method | name | test_region |
|---|---|---|---|---|---|---|---|
| 1024+1 | 36.0 | 0.73 | 1.46 | 0.30 | linear | Intermediate features | Asia |
| 1024+1 | 41.0 | 0.82 | 1.35 | 0.53 | linear | Intermediate features | Europe |
| 1+1 | 36.0 | 0.00 | 1.92 | -0.15 | linear | Baseline prevalence | Asia |
| 1+1 | 41.0 | 0.00 | 2.36 | -0.51 | linear | Baseline prevalence | Europe |
| 18+1 | 36.0 | 0.73 | 1.25 | 0.47 | linear | 18 outputs | Asia |
| 18+1 | 41.0 | 0.86 | 1.04 | 0.71 | linear | 18 outputs | Europe |
| 4+1 | 36.0 | 0.78 | 1.17 | 0.58 | linear | 4 outputs | Asia |
| 4+1 | 41.0 | 0.86 | 1.06 | 0.66 | linear | 4 outputs | Europe |
| 1+1 | 36.0 | 0.77 | 1.15 | 0.57 | linear | lung opacity output | Asia |
| 1+1 | 41.0 | 0.83 | 1.16 | 0.60 | linear | lung opacity output | Europe |
| # params | # test samples | Correlation | MAE | R^2 | method | name | test_region |
|---|---|---|---|---|---|---|---|
| 1024+1 | 36.0 | 0.60 | 1.39 | -0.41 | linear | Intermediate features | Asia |
| 1024+1 | 41.0 | 0.76 | 1.02 | 0.22 | linear | Intermediate features | Europe |
| 1+1 | 36.0 | 0.00 | 1.30 | -0.12 | linear | Baseline prevalence | Asia |
| 1+1 | 41.0 | 0.00 | 1.30 | -0.40 | linear | Baseline prevalence | Europe |
| 18+1 | 36.0 | 0.57 | 1.02 | 0.11 | linear | 18 outputs | Asia |
| 18+1 | 41.0 | 0.76 | 0.79 | 0.48 | linear | 18 outputs | Europe |
| 4+1 | 36.0 | 0.75 | 0.80 | 0.54 | linear | 4 outputs | Asia |
| 4+1 | 41.0 | 0.84 | 0.66 | 0.69 | linear | 4 outputs | Europe |
| 1+1 | 36.0 | 0.74 | 0.83 | 0.53 | linear | lung opacity output | Asia |
| 1+1 | 41.0 | 0.84 | 0.69 | 0.67 | linear | lung opacity output | Europe |
| # params | # test samples | AUPRC | AUROC | method | name | test_region |
|---|---|---|---|---|---|---|
| 1024+1 | {True: 39, False: 2} | 0.95 | 0.50 | logistic | Intermediate features | Asia |
| 1024+1 | {False: 3, True: 7} | 0.70 | 0.50 | logistic | Intermediate features | Americas |
| 1024+1 | {False: 4, True: 3} | 0.60 | 0.75 | logistic | Intermediate features | Oceania |
| 1024+1 | {True: 68, False: 14} | 0.84 | 0.55 | logistic | Intermediate features | Europe |
| 1+1 | {True: 39, False: 2} | 0.95 | 0.50 | logistic | Baseline prevalence | Asia |
| 1+1 | {False: 3, True: 7} | 0.70 | 0.50 | logistic | Baseline prevalence | Americas |
| 1+1 | {False: 4, True: 3} | 0.43 | 0.50 | logistic | Baseline prevalence | Oceania |
| 1+1 | {True: 68, False: 14} | 0.83 | 0.50 | logistic | Baseline prevalence | Europe |
| 18+1 | {True: 39, False: 2} | 0.95 | 0.49 | logistic | 18 outputs | Asia |
| 18+1 | {False: 3, True: 7} | 0.70 | 0.50 | logistic | 18 outputs | Americas |
| 18+1 | {False: 4, True: 3} | 0.60 | 0.75 | logistic | 18 outputs | Oceania |
| 18+1 | {True: 68, False: 14} | 0.86 | 0.59 | logistic | 18 outputs | Europe |
| 4+1 | {True: 39, False: 2} | 0.95 | 0.50 | logistic | 4 outputs | Asia |
| 4+1 | {False: 3, True: 7} | 0.70 | 0.50 | logistic | 4 outputs | Americas |
| 4+1 | {False: 4, True: 3} | 0.43 | 0.50 | logistic | 4 outputs | Oceania |
| 4+1 | {True: 68, False: 14} | 0.83 | 0.52 | logistic | 4 outputs | Europe |
| 1+1 | {True: 39, False: 2} | 0.95 | 0.50 | logistic | lung opacity output | Asia |
| 1+1 | {False: 3, True: 7} | 0.70 | 0.50 | logistic | lung opacity output | Americas |
| 1+1 | {False: 4, True: 3} | 0.43 | 0.50 | logistic | lung opacity output | Oceania |
| 1+1 | {True: 68, False: 14} | 0.83 | 0.51 | logistic | lung opacity output | Europe |
| 5017801 | {True: 39, False: 2} | 0.94 | 0.42 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {True: 39, False: 2} | 0.95 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {True: 39, False: 2} | 0.95 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 3, True: 7} | 0.67 | 0.43 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {False: 3, True: 7} | 0.65 | 0.36 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {False: 3, True: 7} | 0.70 | 0.50 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {False: 4, True: 3} | 0.43 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {False: 4, True: 3} | 0.43 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {False: 4, True: 3} | 0.43 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {True: 68, False: 14} | 0.83 | 0.49 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 68, False: 14} | 0.86 | 0.60 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 68, False: 14} | 0.83 | 0.49 | MLP | Image pixels (MLP) | Europe |
| # params | # test samples | AUPRC | AUROC | method | Features | test_region |
|---|---|---|---|---|---|---|
| 1024+1 | {False: 3, True: 3} | 0.75 | 0.83 | logistic | Intermediate features | Oceania |
| 1024+1 | {True: 72, False: 6} | 0.93 | 0.57 | logistic | Intermediate features | Europe |
| 1+1 | {False: 3, True: 3} | 0.50 | 0.50 | logistic | Baseline prevalence | Oceania |
| 1+1 | {True: 72, False: 6} | 0.92 | 0.50 | logistic | Baseline prevalence | Europe |
| 18+1 | {False: 3, True: 3} | 1.00 | 1.00 | logistic | 18 outputs | Oceania |
| 18+1 | {True: 72, False: 6} | 0.94 | 0.64 | logistic | 18 outputs | Europe |
| 4+1 | {False: 3, True: 3} | 0.50 | 0.50 | logistic | 4 outputs | Oceania |
| 4+1 | {True: 72, False: 6} | 0.93 | 0.54 | logistic | 4 outputs | Europe |
| 1+1 | {False: 3, True: 3} | 0.50 | 0.50 | logistic | lung opacity output | Oceania |
| 1+1 | {True: 72, False: 6} | 0.94 | 0.61 | logistic | lung opacity output | Europe |
| 5017801 | {False: 3, True: 3} | 0.50 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {False: 3, True: 3} | 0.50 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {False: 3, True: 3} | 0.50 | 0.50 | MLP | Image pixels (MLP) | Oceania |
| 5017801 | {True: 72, False: 6} | 0.92 | 0.50 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 72, False: 6} | 0.92 | 0.50 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 72, False: 6} | 0.92 | 0.48 | MLP | Image pixels (MLP) | Europe |
| # params | # test samples | AUPRC | AUROC | method | Features | test_region |
|---|---|---|---|---|---|---|
| 1024+1 | {True: 20, False: 3} | 0.87 | 0.50 | logistic | Intermediate features | Asia |
| 1024+1 | {False: 1, True: 4} | 0.80 | 0.50 | logistic | Intermediate features | Americas |
| 1024+1 | {True: 19, False: 3} | 0.86 | 0.50 | logistic | Intermediate features | Europe |
| 1+1 | {True: 20, False: 3} | 0.87 | 0.50 | logistic | Baseline prevalence | Asia |
| 1+1 | {False: 1, True: 4} | 0.80 | 0.50 | logistic | Baseline prevalence | Americas |
| 1+1 | {True: 19, False: 3} | 0.86 | 0.50 | logistic | Baseline prevalence | Europe |
| 18+1 | {True: 20, False: 3} | 0.86 | 0.45 | logistic | 18 outputs | Asia |
| 18+1 | {False: 1, True: 4} | 0.80 | 0.50 | logistic | 18 outputs | Americas |
| 18+1 | {True: 19, False: 3} | 0.85 | 0.45 | logistic | 18 outputs | Europe |
| 4+1 | {True: 20, False: 3} | 0.87 | 0.50 | logistic | 4 outputs | Asia |
| 4+1 | {False: 1, True: 4} | 0.80 | 0.50 | logistic | 4 outputs | Americas |
| 4+1 | {True: 19, False: 3} | 0.89 | 0.61 | logistic | 4 outputs | Europe |
| 1+1 | {True: 20, False: 3} | 0.87 | 0.50 | logistic | lung opacity output | Asia |
| 1+1 | {False: 1, True: 4} | 0.80 | 0.50 | logistic | lung opacity output | Americas |
| 1+1 | {True: 19, False: 3} | 0.90 | 0.64 | logistic | lung opacity output | Europe |
| 5017801 | {True: 20, False: 3} | 0.87 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {True: 20, False: 3} | 0.87 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {True: 20, False: 3} | 0.87 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 1, True: 4} | 0.80 | 0.50 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {False: 1, True: 4} | 0.80 | 0.50 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {False: 1, True: 4} | 0.80 | 0.50 | MLP | Image pixels (MLP) | Americas |
| 5017801 | {True: 19, False: 3} | 0.86 | 0.50 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 19, False: 3} | 0.86 | 0.50 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {True: 19, False: 3} | 0.86 | 0.50 | MLP | Image pixels (MLP) | Europe |
| # params | # test samples | AUPRC | AUROC | method | Features | test_region |
|---|---|---|---|---|---|---|
| 1024+1 | {False: 9, True: 1} | 0.25 | 0.83 | logistic | Intermediate features | Asia |
| 1024+1 | {False: 15, True: 13} | 0.55 | 0.58 | logistic | Intermediate features | Europe |
| 1+1 | {False: 9, True: 1} | 0.10 | 0.50 | logistic | Baseline prevalence | Asia |
| 1+1 | {False: 15, True: 13} | 0.46 | 0.50 | logistic | Baseline prevalence | Europe |
| 18+1 | {False: 9, True: 1} | 0.33 | 0.89 | logistic | 18 outputs | Asia |
| 18+1 | {False: 15, True: 13} | 0.67 | 0.74 | logistic | 18 outputs | Europe |
| 4+1 | {False: 9, True: 1} | 0.10 | 0.44 | logistic | 4 outputs | Asia |
| 4+1 | {False: 15, True: 13} | 0.61 | 0.66 | logistic | 4 outputs | Europe |
| 1+1 | {False: 9, True: 1} | 0.10 | 0.33 | logistic | lung opacity output | Asia |
| 1+1 | {False: 15, True: 13} | 0.46 | 0.50 | logistic | lung opacity output | Europe |
| 5017801 | {False: 9, True: 1} | 0.11 | 0.56 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 9, True: 1} | 0.10 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 9, True: 1} | 0.10 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 15, True: 13} | 0.46 | 0.30 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {False: 15, True: 13} | 0.46 | 0.27 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {False: 15, True: 13} | 0.46 | 0.27 | MLP | Image pixels (MLP) | Europe |
| # params | # test samples | AUPRC | AUROC | method | Features | test_region |
|---|---|---|---|---|---|---|
| 1024+1 | {False: 10, True: 2} | 0.25 | 0.65 | logistic | Intermediate features | Asia |
| 1024+1 | {False: 8, True: 7} | 0.54 | 0.57 | logistic | Intermediate features | Europe |
| 1+1 | {False: 10, True: 2} | 0.17 | 0.50 | logistic | Baseline prevalence | Asia |
| 1+1 | {False: 8, True: 7} | 0.47 | 0.50 | logistic | Baseline prevalence | Europe |
| 18+1 | {False: 10, True: 2} | 0.17 | 0.30 | logistic | 18 outputs | Asia |
| 18+1 | {False: 8, True: 7} | 0.53 | 0.61 | logistic | 18 outputs | Europe |
| 4+1 | {False: 10, True: 2} | 0.17 | 0.45 | logistic | 4 outputs | Asia |
| 4+1 | {False: 8, True: 7} | 0.45 | 0.45 | logistic | 4 outputs | Europe |
| 1+1 | {False: 10, True: 2} | 0.17 | 0.50 | logistic | lung opacity output | Asia |
| 1+1 | {False: 8, True: 7} | 0.47 | 0.50 | logistic | lung opacity output | Europe |
| 5017801 | {False: 10, True: 2} | 0.17 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 10, True: 2} | 0.17 | 0.50 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 10, True: 2} | 0.18 | 0.55 | MLP | Image pixels (MLP) | Asia |
| 5017801 | {False: 8, True: 7} | 0.50 | 0.56 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {False: 8, True: 7} | 0.47 | 0.38 | MLP | Image pixels (MLP) | Europe |
| 5017801 | {False: 8, True: 7} | 0.47 | 0.50 | MLP | Image pixels (MLP) | Europe |

