1 Introduction
Over time, object detection has firmed its position as a fundamental task in computer vision (Lin et al., 2014; Shao et al., 2019; Geiger et al., 2012). In medical imaging, however, object detection remains heavily under-explored, as the primary focus lies on the dominant discipline of semantic segmentation. This can be mostly traced back to the fact that many clinically relevant tasks require voxel-wise predictions. Object detection, on the other hand, yields object-specific bounding boxes as output. Even though these bounding boxes themselves are of relevancy for object-level diagnostic decision-making (Baumgartner et al., 2021), efficient medical image retrieval and sorting (Criminisi et al., 2010), streamlining complex medical workflows (Gauriau et al., 2015), and robust quantification (Tong et al., 2019), they can be additionally utilized to increase the performance of many medically relevant downstream tasks, such as semantic segmentation (Navarro et al., 2022; Liang et al., 2019), image registration (Samarakoon et al., 2017), or lesions detection (Mamani et al., 2017). In this context, the bounding boxes generated by the preliminary detection step allow the downstream task to focus solely on regions that are likely to contain relevant information, which not only introduces a strong inductive bias but also improves the computation efficiency and unifies the sizes of regions for further analysis. In the specific case of semantic segmentation, the estimated bounding boxes are utilized to crop organ-wise RoIs from the original CT images. Subsequently, these organ-wise RoIs are used to train organ-specific segmentation networks allowing the individual segmentation networks to solely learn highly specialized organ-specific feature representations. This results in a binary instead of a multi-class semantic segmentation problem. However, since this hypothesis only holds if the detection algorithm yields precise results, special attention must be paid towards the development of medical detection algorithms.
Currently, most state-of-the-art architectures operating on 2D natural images exploit the global relation modeling capability of Vision Transformers. These Vision Transformers can generally be divided into two categories, namely Vision Transformer Backbones and Detection Transformers. While Detection Transformers represent complete end-to-end object detection pipelines with an encoder-decoder structure similar to the original machine translation Transformer (Vaswani et al., 2017), Vision Transformer Backbones solely utilize the Transformer’s encoder for feature refinement. To obtain competitive results, object detectors typically combine variants of Vision Transformer Backbones with two-stage (Ren et al., 2015) or multi-stage approaches (Chen et al., 2019) based on convolutional neural networks (CNN).
Although the success of Vision Transformer Backbones has shown that the global relation modeling capability introduced by the attention operation is clearly beneficial, the novel concept of Detection Transformers was unable to produce competitive performances and, therefore, mostly seen as an alternate take on object detection, particularly suitable for set prediction. However, as time evolved, modifications (Zhu et al., 2021; Li et al., 2022; Liu et al., 2022a) of the original Detection Transformer (Carion et al., 2020) narrowed the performance gap to their highly optimized and intensively researched CNN-based counterparts until a Detection Transformer, called DINO (Zhang et al., 2022), was finally able to achieve state-of-the-art results on the COCO benchmark.
Despite the successful employment of Detection Transformers for computer vision tasks processing 2D natural images, the feasibility of adapting Detection Transformers for medical detection tasks remains largely unexplored. The faltering progress of Detection Transformers in the medical domain can be mainly attributed to the absence of large-scale annotated datasets, which are crucial for the successful employment of Detection Transformers (Dosovitskiy et al., 2020). This is because their attention modules require long training schedules to learn sparse, focused attention weights due to their shortage of inductive biases compared to the convolutional operation. In this regard, large-scale datasets are essential to avoid overfitting. Therefore, current medical object detectors still rely predominantly on CNN-based approaches.
In order to accelerate the advent of Detection Transformers to the medical domain, systematic research focusing on their adaption and thus overcoming the limitations imposed by small-scale datasets has to be conducted. Many clinically relevant detection tasks such as organs-at-risk or vertebrae detection rely on a well-defined anatomical field of view (FoV) (Schoppe et al., 2020). This implies that approximate relative and absolute positions of anatomical structures of interest remain consistent throughout the whole dataset. For example, the right lung always appears above the liver in the top right region. We argue that Detection Transformers should bear an immense performance potential for these well-defined FoV anatomical structure detection tasks by presenting three major arguments:
- 1.
The Detection Transformers’ learned positional embeddings, also called query embeddings, should be especially beneficial in the context of the positional consistency assumption of well-defined FoV detection tasks.
- 2.
The concept of relation modeling inherent to Detection Transformers should allow the model to capture relative positional inter-dependencies among anatomical structures via self-attention among object queries.
- 3.
Visualization of attention weights allows for exceptional and accessible explainability of results, which is a crucial aspect of explainable artificial intelligence in medicine.
Therefore, this work aims to pave the path for Detection Transformers, solving the task of 3D anatomical structure detection. We propose a novel Detection Transformer, dubbed Focused Decoder, that alleviates the need for large-scale annotated datasets by reducing the complexity of the 3D detection task. To this end, Focused Decoder builds upon the positional consistency assumption of well-defined FoV detection tasks by exploiting anatomical constraints provided by an anatomical region atlas. Focused Decoder utilizes these anatomical constraints to restrict the cross-attention’s FoV and reintroduce the well-known concept of anchor boxes in the form of query anchors. We prove that Focused Decoder drastically outperforms two Detection Transformer baselines (Carion et al., 2020; Zhu et al., 2021) and performs comparably to the CNN-based detector RetinaNet (Jaeger et al., 2020) while having a significantly lower amount of trainable parameters and increased explainability of results. We summarize our contribution as follows:
- •
To the best of our knowledge, we are the first to successfully leverage 3D Detection Transformers for 3D anatomical structure detection.
- •
We adapt two prominent 2D Detection Transformers, DETR and Deformable DETR, for 3D anatomical structure detection.
- •
We introduce Focused Decoder, a novel Detection Transformer producing strong detection results for well-defined FoV detection tasks by exploiting anatomical constraints provided by an anatomical region atlas to deploy query anchors and restrict the cross-attention’s FoV.
- •
We identify attention weights as a crucial aspect of explainable artificial intelligence in medicine and demonstrate that Focused Decoder improves the explainability of results.
- •
We compare performances of DETR, Deformable DETR, and Focused Decoder with CNN-based approaches represented by RetinaNet on two publicly available computed tomography (CT) datasets and ensure direct comparability of results.
2 Related Works
Since Focused Decoder builds upon prior work, we discuss related work on 2D Detection Transformers processing natural images followed by 3D medical detection algorithms. We pay special attention to detectors featured in this work.
2.1 2D Detection Transformers
DETR (Carion et al., 2020) laid the foundations for Detection Transformers. DETR adopts the machine translation Transformer’s (Vaswani et al., 2017) encoder-decoder architecture to create a streamlined end-to-end object detection pipeline. However, in contrast to the machine translation Transformer, DETR predicts the final set of bounding boxes in parallel rather than in an autoregressive manner. DETR employs a ResNet-based backbone to generate a low resolution representation of the input image, which is subsequently flattened and refined in Transformer encoder blocks. The refined input sequence is in the next step forwarded to the decoder’s cross-attention module to refine a set of object queries. After the object queries have been refined in Transformer decoder blocks, they are fed into a classification and a bounding box regression head, resulting in predicted bounding boxes and class scores. During training, a Hungarian algorithm enforces one-to-one matches, which are particularly suitable for set prediction tasks, based on a set of weighted criteria. Although DETR’s simple design achieves promising 2D detection results on par with a Faster R-CNN baseline, it suffers from high computational complexity, low performance on small objects, and slow convergence.
Deformable DETR (Zhu et al., 2021) improved DETR’s detection performance while simultaneously reducing the model’s computational complexity and training time by introducing the deformable attention mechanism. The concept of deformable attention has been derived from the concept of deformable convolution (Dai et al., 2017), which increases the modeling ability of CNNs by leveraging learned sampling offsets from the original grid sampling locations. The deformable attention module utilizes the learned sparse offset sampling strategy introduced by deformable convolutions by allowing an object query to solely attend to a small fixed set of sampled key points and combines it with the relation modeling ability of the attention operation.
It is worth mentioning that many Detection Transformer variants tried to improve DETR’s initial concept over time. For example, Efficient DETR (Yao et al., 2021) eliminated DETR’s need for an iterative object query refinement process, Conditional DETR (Meng et al., 2021) introduced a conditional cross-attention module, DAB-DETR (Liu et al., 2022a) updated the object query formulation to represent stronger spatial priors, DN-DETR (Li et al., 2022) presented a denoising training strategy, and DINO (Zhang et al., 2022) combined and improved important aspects like denoising training, query initialization, and box prediction.
2.2 3D Medical Detection
Even though 3D medical detection is a longstanding topic in medical image analysis (Criminisi et al., 2009, 2010), most research currently focuses on the more dominant discipline of semantic segmentation (Isensee et al., 2021; Navarro et al., 2021, 2019). This is because most clinically relevant tasks require voxel-vise predictions. Therefore, prior work on deep learning-based 3D medical detection remains relatively limited.
2.2.1 CNN-Based Detectors
Apart from individual experiments with different detectors, such as 3D Faster R-CNN (Xu et al., 2019), most research focused on the CNN-based detector Retina U-Net (Jaeger et al., 2020; Baumgartner et al., 2021). Retina U-Net builds upon the one-stage detector RetinaNet (Lin et al., 2017) and modifies its architecture for 3D medical detection. Retina U-Net’s main contribution is represented by the introduction of a segmentation proxy task. The segmentation proxy task allows the use of voxel-level annotations present in most medical datasets as an additional highly detailed supervisory signal. For detecting objects at different scales, Retina U-Net’s backbone, given by a modified feature pyramid network (FPN), forwards multi-level feature maps to the bounding box regression and classification head networks. Since each voxel of these multi-level feature maps is assigned to a set of 27 level-specific anchor boxes varying in scale, the bounding box regression head predicts position and size offsets, while the classification head predicts their corresponding class scores. As Retina U-Net, therefore, represents a dense one-stage detection scheme, it utilizes non-maximum suppression to reduce duplicates during inference. Retina U-Net is also featured in the automated medical object detection pipeline nnDetection (Baumgartner et al., 2021). Following nnU-Net’s (Isensee et al., 2021) agenda, nnDetection adapts itself without any manual intervention to arbitrary medical detection problems while achieving results on par with or superior to the state-of-the-art. Furthermore, SwinFPN (Wittmann et al., 2022) experimented with Vision Transformer Backbones by incorporating 3D Swin Transformer blocks (Liu et al., 2022b) in Retina U-Net’s architecture, following recent trends in medical semantic segmentation (Hatamizadeh et al., 2022b, a).
2.2.2 Detection Transformers
Although some studies experimented with Detection Transformers operating on 2D medical data (Prangemeier et al., 2020; Shen et al., 2021), only a few attempts tried to adapt them for 3D medical detection tasks. Spine-Transformer (Tao et al., 2022), for example, leverages DETR for sphere-based vertebrae detection. To this end, Spine-Transformer augments DETR with skip connections and additional learnable positional encodings. In contrast to Focused Decoder, however, Spine-Transformer’s concept relies on data of arbitrary FoV. Relationformer (Shit et al., 2022) successfully utilizes Deformable DETR to detect small-scale blood vessels from 3D voxel-level segmentations. The authors introduce an additional relation-token and demonstrate that Relationformer achieves state-of-the-art performances for multi-domain image-to-graph generation tasks, such as blood vessel graph generation.
3 Methods
This section builds our rationale for the proposed Detection Transformer Focused Decoder. Focused Decoder explicitly takes advantage of the fact that approximate relative and absolute positions of labeled anatomical structures contained in datasets of well-defined FoV are consistent, which we try to ensure in an abundant and necessary preprocessing step. We, therefore, draw parallels to the well-known concept of anatomical atlases (Hohne et al., 1992). Based on this positional consistency assumption, we first determine for each dataset FoV-specific anatomical region atlases containing regions of interest (RoI) that comprise labeled anatomical structures. Subsequently, we place in each structure- or class-specific RoI uniformly spaced query anchors and assign a dedicated object query to each of them, resulting in two levels of abstraction. Therefore, Focused Decoder’s object queries are not only assigned to individual RoIs but also to query anchors located inside RoIs. This allows Focused Decoder to restrict the object queries’ FoV for cross-attention to solely voxel within their respective RoI, simplify matching during training and inference, and overcome patient-to-patient variability by generating diverse class-specific predictions enforced by query anchors.
In the following, we introduce the FPN feature extraction backbone and the atlas generation process. Finally, we present the novel concept of query anchors and essential aspects of Focused Decoder, including its architecture, the focused cross-attention module, the concept of relative offset prediction, its matching strategy, and its loss function.
3.1 FPN Feature Extraction Backbone
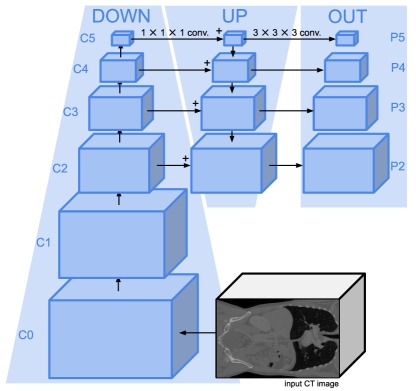
Following DETR, Focused Decoder relies on a feature extraction backbone to create a lower resolution representation of the input CT image , which mitigates the high computational complexity of the attention operation. This feature extraction backbone is depicted in Fig. 1 and is given, similar to Retina U-Net, by an FPN. It should be mentioned that although the feature extraction backbone serves as a CNN-based feature encoder, we refrain from referring to it as an encoder to avoid confusion between the Transformer’s encoder and the FPN. The FPN consists of a CNN-based down-sampling branch (down), an up-sampling branch (up), and a final output projection (out). The up-sampling branch incorporates the down-sampled multi-level feature maps , where , via lateral connections based on convolutions and combines them with up-sampled feature maps of earlier stages. A final output projection acts as an additional refinement stage, fixing the channel dimension to . Therefore, the FPN outputs a set of refined multi-level feature maps , where , encoding semantically strong information by leveraging the top-down inverted high- to low-level information flow. Even though Focused Decoder solely processes the feature map , we require multi-level feature maps for our experiments. This is because our experiments rely on the same feature extraction backbone to ensure maximum comparability of results (see Fig. 4 and Table 5).
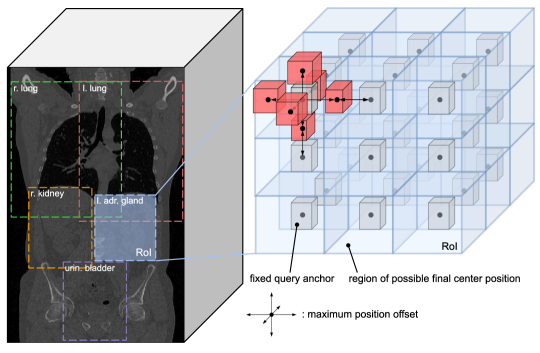
3.2 Atlas Generation
In this paper, we generate custom atlases and refer to them as anatomical region atlases. Relying on the positional consistency assumption, we determine class-specific RoIs, describing the minimum volumes in which all instances of a particular anatomical structure are located. A small subset of RoIs is shown in Fig. 2 (left). Additionally, we estimate for each labeled anatomical structure the median, minimum, and maximum bounding box size, which will be necessary for the query anchor generation process and the concept of relative offset prediction. Given that the test set should only be used to estimate the final performance of the model, we estimate RoIs and bounding box sizes solely based on instances contained in the training and validation sets.
Since medical CT datasets typically contain different labeled anatomical structures, FoVs are inconsistent across datasets. Therefore, we generate dataset-specific anatomical region atlases. In general, however, existing anatomical atlases could be adjusted to fit the datasets at use based on a few anatomical landmarks (Potesil et al., 2013; Xu et al., 2016).
3.3 Query Anchor Generation
Due to dataset-specific inconsistencies and the substantial amount of patient-to-patient variability with regard to positions and sizes of anatomical structures, it is often difficult to ensure that datasets consist of images of exactly the same FoV. This is reflected in the fact that, on average, RoIs occupy 25 times larger volumes than anatomical structures. Therefore, Focused Decoder further subdivides RoIs by assigning to each class-specific RoI in our anatomical region atlas 27 fixed class-specific query anchors in the format . We denote query anchors by . While the spatial locations of query anchors are defined by uniformly spaced positions in their corresponding RoIs, their sizes are governed by the class-specific median bounding box sizes contained in the anatomical region atlas. Generated query anchors associated with the left adrenal gland’s RoI are shown in detail in Fig. 2 (right).
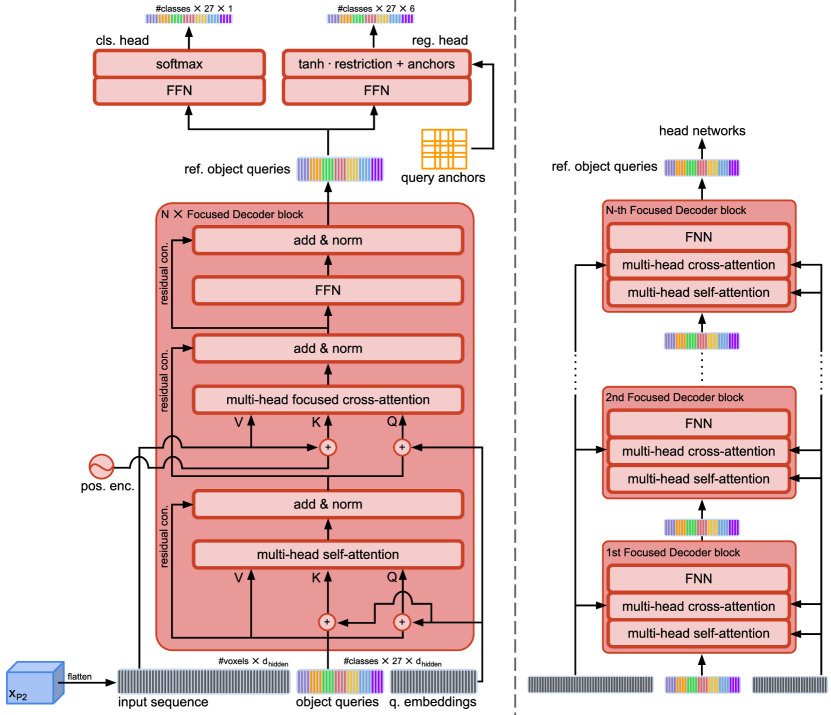
3.4 Focused Decoder
3.4.1 Architecture
The architecture of Focused Decoder and its iterative refinement process are shown in Fig. 3. Focused Decoder represents a lightweight Detection Transformer consisting solely of decoder blocks, omitting the Transformer’s encoder completely. The Transformer’s encoder has the task of modeling relations between elements of the input sequence via its self-attention module. In 3D, this task is highly complex due to long input sequences that arise from flattening high resolution 3D feature maps. Therefore, we argue that omitting the Transformer’s encoder is a sound design choice when training on small-scale medical datasets. Following this hypothesis, Focused Decoder forwards the input sequence given by the flattened feature map directly to the decoder.
Focused Decoder iteratively refines a set of object queries in stacked Focused Decoder blocks (see Fig. 3 (right)). We assign an individual object query to each generated query anchor, which results in , containing class-specific object queries. By assigning object queries to unique spatial locations in the form of query anchors, we demystify their associated learned positional embeddings, called query embeddings, and encourage diverse class-specific predictions, overcoming the issue of patient-to-patient variability. Besides the focused cross-attention module, the general structure of the Focused Decoder block remains similar to DETR’s decoder. The main components of the Focused Decoder block (see Fig. 3 (left)) are represented by a self-attention module followed by a focused cross-attention module and a two-layer feedforward network (FFN), which utilizes a ReLU non-linearity in between its two linear layers. The FFN’s in- and output dimensions correspond to the object queries’ . Its hidden dimension is represented by . While the self-attention module aims to encode strong positional inter-dependencies among object queries, the focused cross-attention module matches the input sequence to object queries and thus regulates the influence of individual feature map voxels for prediction via attention. Subsequently, the FFN facilitates richer feature representations. We employ additional residual connections and layer normalization operations to increase gradient flow.
The classification and bounding box regression head networks are attached to the last Focused Decoder block and process the refined object queries (see Fig. 3 (left)). Given that object queries are already preassigned to specific classes, the classification head predicts class-specific confidence scores, resulting in a binary classification task. The classification head is given by a simple linear layer, whereas the bounding box regression head is represented by a more complex three-layer FFN, which predicts relative position and size offsets with regard to query anchors. The hidden dimension of the bounding box regression head corresponds to the object queries’ . Eventually, we combine these relative offset predictions with their corresponding query anchors and return the predicted bounding boxes together with the class-specific confidence scores, generating overall 27 candidate predictions per class.
Since the attention operation is known to be permutation invariant, valuable information about the spatial location of voxels would be dismissed. To tackle this issue, a positional encoding consisting of sine and cosine functions of different frequencies tries to encode information regarding the absolute position of voxels directly in the input sequence.
3.4.2 Focused Cross-Attention
At the core of Focused Decoder lies the focused cross-attention module forcing class-specific object queries to focus primarily on the structures of interest and their close proximity contained in RoIs of our anatomical region atlas. To this end, the focused cross-attention module leverages an attention mask to restrict the object queries’ FoV for cross-attention. This restriction of attention drastically simplifies the relation modeling task, as we solely have to learn dependencies between object queries and relevant voxels located in their respective RoIs. Equation (1) demonstrates the concept of masked attention, exploited in the focused cross-attention module.
| (1) |
Here, the key and value sequences and are derived from the input sequence, while the object queries contribute to the query sequence . Masking is done by adding the attention mask onto the raw attention weights . The attention mask contains the value for voxels outside and the value for voxels inside the object queries’ respective RoIs. By adding , we nullify the importance of attention weights outside of RoIs for prediction. This is because attention weights corresponding to the value end up in extremely flat areas of the softmax function, which in turn kills gradient flow.
3.4.3 Relative Offset Prediction
Focused Decoder leverages query anchors to take advantage of the concept of relative offset prediction. To this end, we extend the bounding box regression head with a tanh nonlinearity, allowing object queries to modify their query anchors’ fixed center positions and sizes for prediction (see Fig. 3 (left)). However, these modifications are restricted. To counteract overlap and facilitate diverse predictions, center position offsets are restricted to a maximum value, resulting in distinct query-specific regions of possible final center positions covering the RoI completely (see Fig. 2 (right)). In contrast to position offset restrictions, which are derived from the sizes of RoIs, we restrict the allowed size offsets to the minimum and maximum bounding box sizes contained in our anatomical region atlas.
3.4.4 Matching
Since Focused Decoder predicts 27 eligible candidates per class, matching boils down to finding the most suitable candidate among 27. To train the head networks in a meaningful way, we generate dynamic confidence labels, solely relying on the normalized generalized intersection over union (GIoU) between all class-specific query anchors and their corresponding ground truth objects. This results in dynamic confidence labels of 1 for predictions corresponding to query anchors with the highest GIoU per class and dynamic confidence labels of 0 for predictions corresponding to query anchors with the lowest GIoU per class. Based on these dynamic confidence labels, we match predictions with the highest dynamic confidence labels per class to the ground truth objects. We subsequently forward matched predictions and the dynamic confidence labels to the loss function.
3.4.5 Loss Function
Equation (2) expresses Focused Decoder’s general loss function, where corresponds to the number of Focused Decoder blocks. Following Deformable DETR, , , and correspond to 2, 2, and 5.
| (2) |
We utilize a binary cross-entropy loss between predicted confidence scores and dynamic confidence labels determined during matching to facilitate diverse predictions during inference. For bounding box regression, two loss functions, namely a scale-invariant GIoU loss and an loss , are combined. The bounding box regression loss functions solely consider matched predictions and thus only optimize predictions having dynamic confidence labels of 1. Following DETR, we additionally forward predictions generated based on outputs of earlier Focused Decoder blocks (see Fig. 3 (right)) to the final loss function, resulting in an auxiliary loss for intermediate supervision.
3.4.6 Inference
During inference, the class-specific confidence scores predicted by the classification head indicate the most suitable candidates. Therefore, we simply select predictions with the highest confidence scores per class to represent the output.
4 Experiments and Results
In this section, we determine Focused Decoder’s performance for 3D anatomical structure detection and compare it to two Detection Transformer baselines and a RetinaNet variant adopted from the state-of-the-art detection pipeline nnDetection. Subsequently, we demonstrate the excellent explainability of Focused Decoder’s results and critically assess the importance of its design choices.
4.1 Experimental Setup
First, it should be mentioned that comparability of performances is of utmost importance and, due to the lack of 3D anatomical structure detection benchmarks, challenging to achieve. Therefore, we adapt and directly integrate detectors featured in this work into the same detection and training pipeline for a fair and reproducible comparison. For example, all featured detectors were trained until convergence on a single Quadro RTX 8000 GPU using the same AdamW optimizer, step learning rate scheduler, data augmentation techniques, and FPN feature extraction backbone. In addition, we tried to keep the configurations of featured Detection Transformers as similar as possible. We use the same head networks, set to three, to 384, and to 1024. By doing so, we ensure maximal comparability of results. Precise information about training details, hyperparameters, and individual model configurations is available at https://github.com/bwittmann/transoar. To facilitate understanding of our experimental setup, Fig. 4 depicts an overview of all featured detectors. Next, we briefly introduce the Detection Transformer baselines, which we adapted for 3D anatomical structure detection, and our RetinaNet variant.
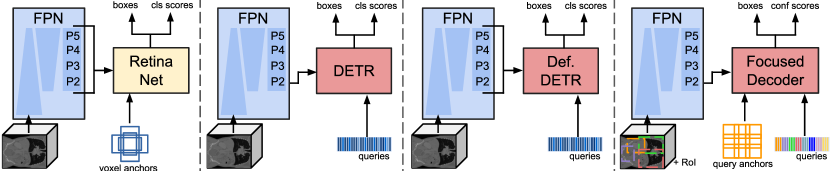
4.1.1 DETR
DETR acts as our first Detection Transformer baseline. Besides some additional minor adjustments, the general structure and configuration of our DETR variant remain essentially unchanged. Following the original DETR’s configuration, we set the number of object queries, which are in contrast to Focused Decoder not preassigned to specific classes, to 100. While the Hungarian algorithm is responsible for matching during training, we return the highest-scoring predictions per class during inference.
4.1.2 Deformable DETR
To evaluate Focused Decoder properly, we introduce Deformable DETR as an additional Detection Transformer baseline. Deformable DETR generates its input sequence by flattening and combining the multi-level feature maps of the stages P2 to P5 into a combined input sequence and refines, similar to DETR, a set of 100 object queries. Deformable DETR employs the same matching strategies during inference and training as DETR.
4.1.3 RetinaNet
As a representative of CNN-based detectors, we adopt Retina U-Net from nnDetection with minor modifications. Since the automated detection pipeline nnDetection developed around Retina U-Net has been shown to deliver state-of-the-art results for numerous medical detection tasks, we extract nnDetection’s generated hyperparameters and refine them via a brief additional hyperparameter search. In addition, we omit Retina U-Net’s segmentation proxy task, converting it to a RetinaNet variant. This not only enforces comparability with other detectors featured in this work but also drastically reduces training times. We justify this decision as experiments111We provide experiments with the segmentation proxy task incorporated in all featured detectors in our GitHub repository. To activate the segmentation proxy task, set flag use_seg_proxy_loss to True in the respective config file. have shown that the segmentation proxy task leads to extremely minor and hence negligible performance benefits for 3D anatomical structure detection.
4.2 Datasets
Due to the lack of 3D anatomical structure detection benchmarks, we conduct experiments on two CT datasets of well-defined FoV that were originally developed for the task of semantic segmentation, namely the VISCERAL anatomy benchmark (Jimenez-del Toro et al., 2016) and the AMOS22 challenge (Ji et al., 2022). We transform their voxel-wise annotations into bounding boxes and class labels in the dataloader. Dataset-specific spatial sizes, approximated resolutions, and sizes of the respective training, validation, and test sets are along with the total number of subjects reported in Table 1.
| Dataset | Size | Resolution (in mm) | Train / Val / Test | # of subjects |
| VISCERAL | 120 / 20 / 12 | 152 | ||
| AMOS22 | 117 / 20 / 18 | 155 |
4.2.1 VISCERAL Anatomy Benchmark
The VISCERAL anatomy benchmark contains segmentations of 20 major anatomical structures. The CT images of the silver corpus subset are used as training data, while the gold corpus subset is split into two halves to create the validation and test sets.
4.2.2 AMOS22 Challenge
The multi-modality abdominal multi-organ segmentation challenge of 2022, short AMOS22, provides CT images with voxel-level annotations of 15 abdominal organs. We divide the CT images of the challenge’s first stage into the training, validation, and test sets.
4.2.3 Preprocessing
We utilize the same preprocessing approach for both datasets. First, the raw CT images and labels are transformed to a uniform orientation represented by the RAS, short for ’right, anterior, superior’, axis code. We subsequently crop CT images to foreground structures. By doing so, we additionally try to ensure datasets consisting of images with a similar FoV, which is necessary to determine meaningful class-specific RoIs. In the next step, the CT images and their labels are resized to a fixed spatial size. This increases fairness and further reduces the detection task’s complexity by compensating for variations in patient body size. To compensate for incompletely labeled CT images, we discard CT images based on a specified label threshold, allowing for a sufficient amount of data while simultaneously ensuring fairly clean datasets. Additionally, we completely omit partially labeled CT images in the test sets to provide a valid performance estimate.
4.3 Metric
The metric commonly used to evaluate object detection algorithms is the mean average precision (mAP). The mAP metric, described in (3), is defined as the mean of a selected subset of average precision (AP) values. We report , which is calculated based on AP values evaluated at IoU thresholds = {0.5, 0.55, …, 0.95}.
| (3) |
To determine detection performance related to structure size, we introduce , , and . To this end, we categorize classes based on the volume occupancy of their median bounding boxes into the subsets S (small), M (medium), and L (large), which rely on the volume occupancy ranges of [0.0%, 0.5%), [0.5%, 5.0%), and [5.0%, 100%], respectively. Subsequently, we reevaluate restricted to solely classes in the individual subsets. The dataset-specific subsets determined based on the above reported volume occupancy ranges are shown in Table 2. It should be mentioned that although both datasets have anatomical structures in common, they might be assigned to different subsets due to varying FoVs. The difference in FoV across both datasets can be observed in Fig. 7 (left).
| VISCERAL | S | pancreas, gall bladder, urinary bladder, trachea, thyroid gland,first lumbar vertebra, adrenal glands |
| M | spleen, aorta, sternum, kidneys, psoas major, rectus abdominis | |
| L | liver, lungs | |
| AMOS | S | esophagus, adrenal glands, prostate/uterus |
| M | spleen, kidneys, gall bladder, aorta, postcava, pancreas, duodenum,urinary bladder | |
| L | liver, stomach |
| Dataset | Model | |||||
| VISCERAL | RetinaNet | 52.8M | 41.37* | 20.99* | 47.02 | 78.74 |
| DETR | 46.5M | 27.35 | 11.32 | 28.10 | 67.88 | |
| Def. DETR | 54.8M | 33.06 | 14.52 | 35.09 | 76.39 | |
| Focused Decoder | 41.8M | 39.22 | 18.33 | 43.59 | 81.70* | |
| AMOS22 | RetinaNet | 52.1M | 30.38 | 16.92* | 32.37 | 44.38 |
| DETR | 43.7M | 16.47 | 4.24 | 15.68 | 32.96 | |
| Def. DETR | 53.4M | 26.59 | 8.93 | 30.32 | 43.08 | |
| Focused Decoder | 42.6M | 29.83 | 10.30 | 34.01 | 47.97* | |
| * denotes statistically significant difference between Focused Decoder and RetinaNet; p-values . | ||||||
4.4 Quantitative Results
Table 3 lists quantitative results of all detectors featured in this work. Essentially, our findings remain consistent across both datasets. Focused Decoder, possessing the least amount of trainable parameters, significantly outperforms the Detection Transformer baselines, namely DETR and Deformable DETR, and performs close to the state-of-the-art detector RetinaNet.
This demonstrates that a naive adaption of 2D Detection Transformers to the medical domain is definitely not sufficient to overcome the problem of data scarcity. However, Focused Decoder’s strong detection performances reveal that reducing the complexity of the relation modeling and thus the 3D detection task alleviates the need for large-scale annotated datasets, converting Detection Transformers into competitive and lightweight (approximately fewer parameters compared to RetinaNet) 3D medical detection algorithms. Focused Decoder not only performs close to or on par with its CNN-based counterpart but also provides the most accurate predictions for larger structures. On the other hand, RetinaNet outperforms Focused Decoder on smaller structures, overcoming the Detection Transformer’s well-known shortcoming of poor small object detection performance, which is subject to open research.
Even though the overall detection performance on the VISCERAL anatomy benchmark turns out to be stronger, Focused Decoder manages to narrow the gap to RetinaNet on the AMOS22 challenge. The generally stronger performances on the VISCERAL anatomy benchmark can be attributed to the absence of symmetrical and easy to detect structures such as the lungs or the rectus abdominis in the AMOS22 challenge. The narrowed performance gap, however, primarily results from the difference in FoV between both datasets. This is because the reduced FoV of the AMOS22 challenge (see Fig. 7 (left)) leads to an artificial increase in structure size, which is most beneficial for Focused Decoder.
To estimate the statistical significance of the performance differences between Focused Decoder and RetinaNet, we determined p-values using the Wilcoxon signed-rank test based on bootstrapped subsets. We indicate statistically significant performance differences in Table 3 (see footnote).
4.5 Explainability of Focused Decoder’s Predictions
Explainability of results is of exceptional importance in the context of medical image analysis. While identifying reasons behind predictions produced by CNN-based detectors is often cumbersome and requires additional engineering susceptible to errors, Focused Decoder’s attention weights facilitate the accessibility of explainable results. Therefore, we particularly probe into intra-anatomical and inter-anatomical explainability by analyzing cross- and self-attention weights.
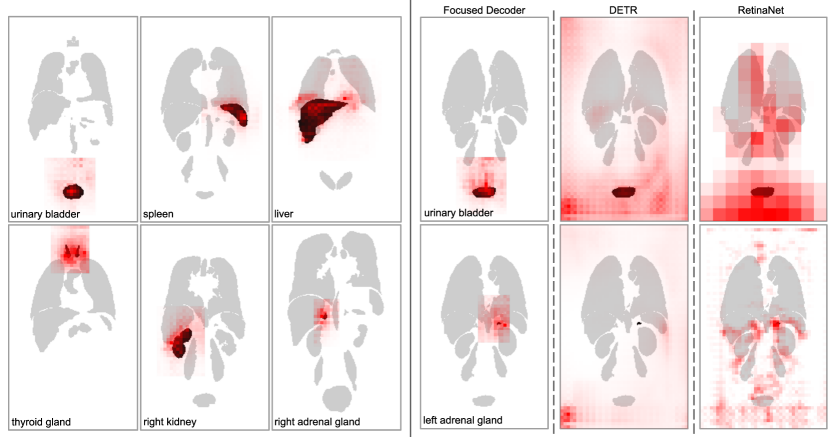
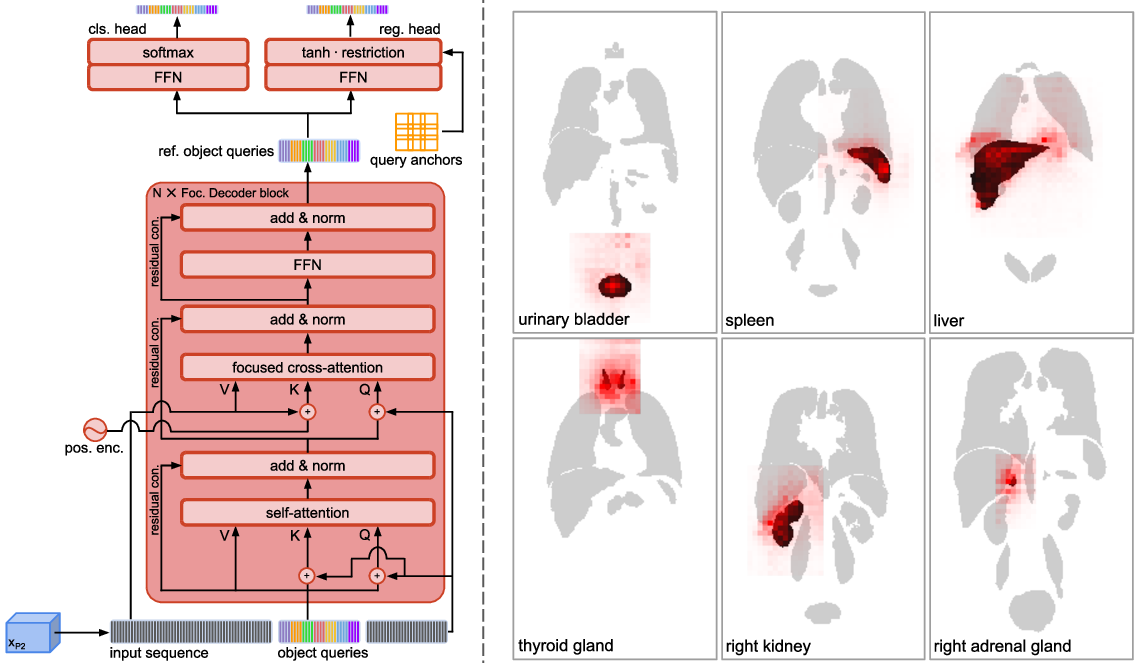
4.5.1 Intra-Anatomical Explainability
Focused Decoder’s cross-attention weights between the input sequence and object queries corresponding to selected anatomical structures are visualized in Fig. 5 (left). Since cross-attention weights indicate the importance of voxels for prediction, one can conclude that Focused Decoder’s predictions primarily rely on structures of interest and the context in their proximity, even for extremely small objects such as the adrenal glands.
To highlight Focused Decoder’s intra-anatomical explainability of results, we additionally compare its cross-attention weights to cross-attention weights generated by DETR and gradient-weighted class activation maps corresponding to RetinaNet’s predictions in Fig. 5 (right). These gradient-weighted activation maps were generated using the established Grad-CAM (Selvaraju et al., 2017) algorithm, backpropagating to the FPN’s output feature maps. Focused Decoder’s cross-attention weights provide the best explainability of results. In contrast, DETR’s cross-attention weights and RetinaNet’s class activations are scattered over large parts of the CT image, complicating explainability and possibly indicating memorization of dataset-specific covariates.
4.5.2 Inter-Anatomical Explainability
While cross-attention weights indicate the importance of voxels for prediction, self-attention weights reveal the inter-dependencies among the class-specific object queries, providing inter-anatomical explainability. We visualize self-attention weights in Fig. 6 to prove the hypothesis that Focused Decoder captures meaningful inter-dependencies among anatomical structures.
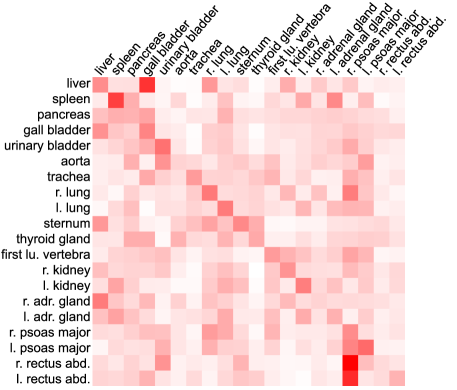
We observe the trend of high inter-dependencies among neighboring structures. Object queries corresponding to the liver (first row), for example, attend primarily to structures in the liver’s proximity, such as the gall bladder, the right lung, the right kidney, and the right psoas major, whereas object queries corresponding to the spleen (second row) attend primarily to the pancreas, the left lung, the left kidney, and the left adrenal gland.
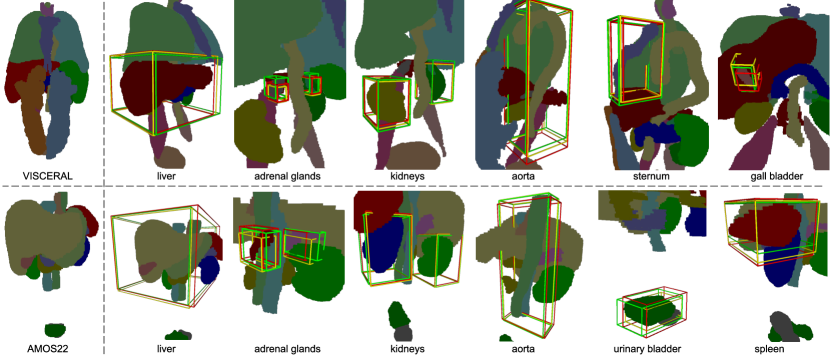
4.6 Qualitative Results
Qualitative results in the form of predicted bounding boxes of the two best-performing architectures, RetinaNet and Focused Decoder, are compared to the ground truth in Fig. 7. One can observe that Focused Decoder exhibits exceptional detection performance on larger structures such as the liver or the aorta, while RetinaNet remains superior in detecting small structures such as the adrenal glands.
4.7 Ablation Studies
In this subsection, we investigate the importance of Focused Decoder’s design choices by conducting detailed ablations on the validation set of the VISCERAL anatomy benchmark. Table 4 displays values produced by Focused Decoder’s base configuration (first row) and three additional configurations (second to fourth row), omitting different design choices.
| Restriction | Anchors | Queries per class | ||
| ✓ | ✓ | 27 | 37.78 | |
| ✓ | 27 | 36.63 | -1.15 | |
| ✓ | 1 | 35.08 | -2.70 | |
| ✓ | 27 | 32.03 | -5.75 |
To evaluate the impact of our proposed query anchors, we completely omit them in the second configuration, resulting in an decrease of 1.15. This proves the importance of demystifying object queries and their associated query embeddings by assigning them to precise spatial locations and thus generating diverse class-specific predictions, overcoming the issue of patient-to-patient variability. The configuration presented in the third row additionally reduces the amount of object queries per class from 27 to one. Based on the reduction of 2.70, we argue that having only one object query per class fails to cover all class-specific object variations and hence leads to performance decreases. Next, we deactivate the focused cross-attention module’s restriction to RoIs in the fourth row. As expected, detection performance drastically diminishes, which is reflected in an delta of -5.75. The result of this ablation study repeatedly demonstrates the necessity of simplifying the relation modeling task to achieve competitive detection performances.
Since the FPN outputs a set of refined multi-level feature maps , we experiment with different input sequences represented by flattened feature maps of different resolutions and report our findings in Table 5.
| Stage | Feature map resolution | ||
| P2 | 37.78 | ||
| P3 | 36.97 | -0.81 | |
| P4 | 33.72 | -4.07 | |
| P5 | 29.96 | -7.82 |
One can observe that feature maps of higher resolution are clearly beneficial for detection performance, as they contain more fine-grained details, which in turn leads to more precise bounding box estimations. However, the performance improvement saturates as the feature map resolutions increase.
Finally, we prove that omitting the Transformer’s encoder leads to more precise detections when the amount of available training data is strictly limited. To this end, we experiment with DETR and Deformable DETR configurations omitting the encoder and report the results in Table 6.
| Model | Encoder | |||
| DETR | 25.94 | 61.49 | 17.46 | |
| DETR | ✓ | 23.32 | 59.11 | 14.42 |
| Def DETR | 30.95 | 67.07 | 24.35 | |
| Def DETR | ✓ | 29.26 | 65.31 | 19.44 |
Evaluation of values presented in Table 6 leads to the conclusion that the Transformer’s encoder is disruptive to detection performance. This supports the hypothesis that the encoder’s intricate relation modeling task would require an extreme amount of training epochs and hence a large amount of annotated data to capture meaningful dependencies among the input sequence elements.
5 Limitations
We would like to particularly highlight the limitations of Focused Decoder’s design, which can be primarily attributed to the assumption of well-defined FoVs. Although this assumption is reasonable to make from a clinical standpoint, it also poses a significant challenge to Focused Decoder’s robustness with regard to CT images of varying FoVs. This is because drastic shifts in FoV may lead to anatomical structures being located partially outside of RoIs, which in turn may hinder object queries to encode meaningful information via our proposed focused cross-attention module. However, it should be emphasized that the FoV of CT images could always be adjusted to fit the FoVs of our datasets by adopting our preprocessing step or based on registration via a few anatomical landmarks.
It is also worth mentioning that Focused Decoder predicts by design for each anatomical structure contained in our anatomical region atlases exactly one final bounding box. Therefore, the complete absence of anatomical structures, for example after radical nephrectomy, would remain unnoticed.
6 Outlook and Conclusion
This work lays the foundations of 3D medial Detection Transformers by introducing Focused Decoder, a lightweight alternative to CNN-based detectors, which not only exhibits exceptional and highly intuitive explainability of results but also demonstrates the best detection performances on large structures. Focused Decoder’s impressive performances on large structures already conclusively indicate the immense potential of Detection Transformers for medical applications.
Based on results from our experiments, we recommend the use of Focused Decoder for anatomical structure detection tasks when the priority lies on the analysis of large structures or the explainability of results. However, we strongly believe that with increasing sizes of annotated medical datasets, Detection Transformers will eventually outperform CNN-based architectures on medical detection tasks in all metrics, resulting in a complete shift from CNN- to Transformer-based architectures.
We encourage future work to further explore Focused Decoder’s parameters, overcome its limitations, and address its inferior performance on small structures by investigating the influence of the fixed FoV of CT images, which results in drastic size differences between anatomical structures (the right lung, for example, occupies magnitudes more space compared to the urinary bladder in CT images of fixed FoV). Future work should, therefore, also focus on exploring approaches operating on uniformly sized RoIs and hence dynamic spatial resolutions. This would in turn lead to uniform structure sizes, possibly allowing Focused Decoder to overcome the issue of relative scale between anatomical structures.
Acknowledgments
This work has been supported in part by the TRABIT under the EU Marie Sklodowska-Curie Program (Grant agreement ID: 765148) and in part by the DCoMEX (Grant agreement ID: 956201). The work of Bjoern Menze was supported by a Helmut-Horten-Foundation Professorship.
Ethical Standards
The work follows appropriate ethical standards in conducting research and writing the manuscript, following all applicable laws and regulations regarding treatment of animals or human subjects.
Conflicts of Interest
We declare we don’t have conflicts of interest.
References
- Baumgartner et al. (2021) Michael Baumgartner, Paul F Jäger, Fabian Isensee, and Klaus H Maier-Hein. nnDetection: A self-configuring method for medical object detection. In Proc. MICCAI, pages 530–539, 2021.
- Carion et al. (2020) Nicolas Carion, Francisco Massa, Gabriel Synnaeve, Nicolas Usunier, Alexander Kirillov, and Sergey Zagoruyko. End-to-end object detection with transformers. In Proc. ECCV, pages 213–229, 2020.
- Chen et al. (2019) Kai Chen, Jiangmiao Pang, Jiaqi Wang, Yu Xiong, Xiaoxiao Li, Shuyang Sun, Wansen Feng, Ziwei Liu, Jianping Shi, Wanli Ouyang, Chen Change Loy, and Dahua Lin. Hybrid task cascade for instance segmentation. In Proc. IEEE/CVF CVPR, pages 4974–4983, 2019.
- Criminisi et al. (2009) Antonio Criminisi, Jamie Shotton, and Stefano Bucciarelli. Decision forests with long-range spatial context for organ localization in CT volumes. In Proc. MICCAI, pages 69–80, 2009.
- Criminisi et al. (2010) Antonio Criminisi, Jamie Shotton, Duncan Robertson, and Ender Konukoglu. Regression forests for efficient anatomy detection and localization in CT studies. In Proc. MCV Workshop, pages 106–117, 2010.
- Dai et al. (2017) Jifeng Dai, Haozhi Qi, Yuwen Xiong, Yi Li, Guodong Zhang, Han Hu, and Yichen Wei. Deformable convolutional networks. In Proc. IEEE ICCV, pages 764–773, 2017.
- Dosovitskiy et al. (2020) Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, and Neil Houlsby. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, 2020.
- Gauriau et al. (2015) Romane Gauriau, Rémi Cuingnet, David Lesage, and Isabelle Bloch. Multi-organ localization with cascaded global-to-local regression and shape prior. MedIA, 23(1):70–83, 2015.
- Geiger et al. (2012) Andreas Geiger, Philip Lenz, and Raquel Urtasun. Are we ready for autonomous driving? the KITTI vision benchmark suite. In Proc. IEEE/CVF CVPR, pages 3354–3361, 2012.
- Hatamizadeh et al. (2022a) Ali Hatamizadeh, Vishwesh Nath, Yucheng Tang, Dong Yang, Holger R Roth, and Daguang Xu. Swin UNETR: Swin transformers for semantic segmentation of brain tumors in mri images. arXiv preprint arXiv:2201.01266, 2022a.
- Hatamizadeh et al. (2022b) Ali Hatamizadeh et al. UNETR: Transformers for 3d medical image segmentation. In Proc. IEEE/CVF WACV, pages 574–584, 2022b.
- Hohne et al. (1992) Karl Heinz Hohne, Michael Bomans, Martin Riemer, Rainer Schubert, Ulf Tiede, and Werner Lierse. A volume-based anatomical atlas. IEEE CGA, 12(04):73–77, 1992.
- Isensee et al. (2021) Fabian Isensee, Paul F Jaeger, Simon AA Kohl, Jens Petersen, and Klaus H Maier-Hein. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2):203–211, 2021.
- Jaeger et al. (2020) Paul F Jaeger, Simon AA Kohl, Sebastian Bickelhaupt, Fabian Isensee, Tristan Anselm Kuder, Heinz-Peter Schlemmer, and Klaus H Maier-Hein. Retina U-Net: embarrassingly simple exploitation of segmentation supervision for medical object detection. In Proc ML4H, pages 171–183. PMLR, 2020.
- Ji et al. (2022) Yuanfeng Ji, Haotian Bai, Jie Yang, Chongjian Ge, Ye Zhu, Ruimao Zhang, Zhen Li, Lingyan Zhang, Wanling Ma, Xiang Wan, et al. AMOS: a large-scale abdominal multi-organ benchmark for versatile medical image segmentation. arXiv preprint arXiv:2206.08023, 2022.
- Jimenez-del Toro et al. (2016) Oscar Jimenez-del Toro, Henning Müller, Markus Krenn, Katharina Gruenberg, Abdel Aziz Taha, Marianne Winterstein, Ivan Eggel, Antonio Foncubierta-Rodríguez, Orcun Goksel, András Jakab, et al. Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks. IEEE TMI, 35(11):2459–2475, 2016.
- Li et al. (2022) Feng Li, Hao Zhang, Shilong Liu, Jian Guo, Lionel M Ni, and Lei Zhang. Dn-DETR: Accelerate DETR training by introducing query denoising. In Proc. IEEE/CVF CVPR, pages 13619–13627, 2022.
- Liang et al. (2019) Shujun Liang, Fan Tang, Xia Huang, Kaifan Yang, Tao Zhong, Runyue Hu, Shangqing Liu, Xinrui Yuan, and Yu Zhang. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. European radiology, 29(4):1961–1967, 2019.
- Lin et al. (2014) Tsung-Yi Lin, Michael Maire, Serge Belongie, James Hays, Pietro Perona, Deva Ramanan, Piotr Dollár, and C Lawrence Zitnick. Microsoft COCO: common objects in context. In Proc. ECCV, pages 740–755, 2014.
- Lin et al. (2017) Tsung-Yi Lin, Priya Goyal, Ross Girshick, Kaiming He, and Piotr Dollár. Focal loss for dense object detection. In Proc. IEEE ICCV, pages 2980–2988, 2017.
- Liu et al. (2022a) Shilong Liu, Feng Li, Hao Zhang, Xiao Yang, Xianbiao Qi, Hang Su, Jun Zhu, and Lei Zhang. DAB-DETR: Dynamic anchor boxes are better queries for DETR. arXiv preprint arXiv:2201.12329, 2022a.
- Liu et al. (2022b) Ze Liu, Jia Ning, Yue Cao, Yixuan Wei, Zheng Zhang, Stephen Lin, and Han Hu. Video Swin transformer. In Proc. IEEE/CVF CVPR, pages 3202–3211, 2022b.
- Mamani et al. (2017) Gabriel Efrain Humpire Mamani, Arnaud Arindra Adiyoso Setio, Bram van Ginneken, and Colin Jacobs. Organ detection in thorax abdomen CT using multi-label convolutional neural networks. In Proc Medical Imaging: Computer-Aided Diagnosis, pages 287–292, 2017.
- Meng et al. (2021) Depu Meng, Xiaokang Chen, Zejia Fan, Gang Zeng, Houqiang Li, Yuhui Yuan, Lei Sun, and Jingdong Wang. Conditional DETR for fast training convergence. In Proc. IEEE/CVF CVPR, pages 3651–3660, 2021.
- Navarro et al. (2019) Fernando Navarro, Suprosanna Shit, Ivan Ezhov, Johannes Paetzold, Andrei Gafita, Jan C Peeken, Stephanie E Combs, and Bjoern H Menze. Shape-aware complementary-task learning for multi-organ segmentation. In Proc. MLMI, pages 620–627, 2019.
- Navarro et al. (2021) Fernando Navarro, Christopher Watanabe, Suprosanna Shit, Anjany Sekuboyina, Jan C Peeken, Stephanie E Combs, and Bjoern H Menze. Evaluating the robustness of self-supervised learning in medical imaging. arXiv preprint arXiv:2105.06986, 2021.
- Navarro et al. (2022) Fernando Navarro, Guido Sasahara, Suprosanna Shit, Ivan Ezhov, Jan C Peeken, Stephanie E Combs, and Bjoern H Menze. A unified 3D framework for organs at risk localization and segmentation for radiation therapy planning. arXiv preprint arXiv:2203.00624, 2022.
- Potesil et al. (2013) Vaclav Potesil, Timor Kadir, and Michael Brady. Learning new parts for landmark localization in whole-body CT scans. IEEE TMI, 33(4):836–848, 2013.
- Prangemeier et al. (2020) Tim Prangemeier, Christoph Reich, and Heinz Koeppl. Attention-based transformers for instance segmentation of cells in microstructures. In Proc. IEEE BIBM, pages 700–707, 2020.
- Ren et al. (2015) Shaoqing Ren, Kaiming He, Ross Girshick, and Jian Sun. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proc. NeurIPS, pages 91–99, 2015.
- Samarakoon et al. (2017) Prasad N Samarakoon, Emmanuel Promayon, and Celine Fouard. Light random regression forests for automatic multi-organ localization in CT images. In Proc IEEE ISBI, pages 371–374, 2017.
- Schoppe et al. (2020) Oliver Schoppe, Chenchen Pan, Javier Coronel, Hongcheng Mai, Zhouyi Rong, Mihail Ivilinov Todorov, Annemarie Müskes, Fernando Navarro, Hongwei Li, Ali Ertürk, et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nature communications, 11(1):1–14, 2020.
- Selvaraju et al. (2017) Ramprasaath R Selvaraju, Michael Cogswell, Abhishek Das, Ramakrishna Vedantam, Devi Parikh, and Dhruv Batra. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proc. IEEE ICCV, pages 618–626, 2017.
- Shao et al. (2019) Shuai Shao, Zeming Li, Tianyuan Zhang, Chao Peng, Gang Yu, Xiangyu Zhang, Jing Li, and Jian Sun. Objects365: A large-scale, high-quality dataset for object detection. In Proc. IEEE/CVF CVPR, pages 8430–8439, 2019.
- Shen et al. (2021) Zhiqiang Shen, Rongda Fu, Chaonan Lin, and Shaohua Zheng. COTR: convolution in transformer network for end to end polyp detection. In Proc. INFOCOM, pages 1757–1761, 2021.
- Shit et al. (2022) Suprosanna Shit, Rajat Koner, Bastian Wittmann, Johannes Paetzold, Ivan Ezhov, Hongwei Li, Jiazhen Pan, Sahand Sharifzadeh, Georgios Kaissis, Volker Tresp, et al. Relationformer: A unified framework for image-to-graph generation. arXiv preprint arXiv:2203.10202, 2022.
- Tao et al. (2022) Rong Tao, Wenyong Liu, and Guoyan Zheng. Spine-transformers: Vertebra labeling and segmentation in arbitrary field-of-view spine CTs via 3D transformers. MedIA, 75:102258, 2022.
- Tong et al. (2019) Yubing Tong, Jayaram K Udupa, Dewey Odhner, Caiyun Wu, Stephen J Schuster, and Drew A Torigian. Disease quantification on PET/CT images without explicit object delineation. MedIA, 51:169–183, 2019.
- Vaswani et al. (2017) Ashish Vaswani, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N Gomez, Łukasz Kaiser, and Illia Polosukhin. Attention is all you need. In Proc. NeurIPS, pages 5998–6008, 2017.
- Wittmann et al. (2022) Bastian Wittmann, Suprosanna Shit, Fernando Navarro, Jan C Peeken, Stephanie E Combs, and Bjoern H Menze. SwinFPN: Leveraging vision transformers for 3d organs-at-risk detection. In Proc. MIDL, 2022.
- Xu et al. (2019) Xuanang Xu, Fugen Zhou, Bo Liu, Dongshan Fu, and Xiangzhi Bai. Efficient multiple organ localization in CT image using 3D region proposal network. IEEE TMI, 38(8):1885–1898, 2019.
- Xu et al. (2016) Zhoubing Xu, Christopher P Lee, Mattias P Heinrich, Marc Modat, Daniel Rueckert, Sebastien Ourselin, Richard G Abramson, and Bennett A Landman. Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE TBME, 63(8):1563–1572, 2016.
- Yao et al. (2021) Zhuyu Yao, Jiangbo Ai, Boxun Li, and Chi Zhang. Efficient DETR: improving end-to-end object detector with dense prior. arXiv preprint arXiv:2104.01318, 2021.
- Zhang et al. (2022) Hao Zhang, Feng Li, Shilong Liu, Lei Zhang, Hang Su, Jun Zhu, Lionel M Ni, and Heung-Yeung Shum. Dino: DETR with improved denoising anchor boxes for end-to-end object detection. arXiv preprint arXiv:2203.03605, 2022.
- Zhu et al. (2021) Xizhou Zhu, Weijie Su, Lewei Lu, Bin Li, Xiaogang Wang, and Jifeng Dai. Deformable DETR: Deformable transformers for end-to-end object detection. In Proc. ICLR, 2021.