1 Introduction
In cancer staging, the N-staging component of the Classification of Malignant Tumors (TNM) classification system provides insights into the presence of metastases in regional lymph nodes. Accurate identification of metastatic lymph nodes poses a challenge for diagnosis through CT imaging alone due to minimal contrast differences with surrounding tissue and strong variations in size, shape, number, and location of lymph nodes. PET/CT scans serve as a gold standard to assess functional parameters, that is metabolic activity and, consequently, lymph node malignancy. When PET scans are unavailable due to high examination costs and radioactive exposure to the patient, only CT images are used. In CT, in contrast to PET, only morphological factors such as the lymph node size can be evaluated. The Response Evaluation Criteria in Solid Tumors (RECIST) introduced by Eisenhauer et al. (2009) is commonly used in this context. It defines a lymph node to be pathological if its short-axis diameter exceeds 10 mm in axial plane. With the above-mentioned challenges of distinguishing lymph nodes from surrounding soft tissue and its highly resource-intensive manual assessment, there arises a need for robust and performant algorithms to tackle the task of detection/segmentation of cancerous lymph node for lymph node staging without human interaction. Automated segmentation of pathological lymph nodes facilitates tumor staging based on both PET/CT or CT only, and subsequently supports decision-making regarding the necessity of surgery and further treatment.
The LNQ 2023 challenge hosted at MICCAI 2023 aims to provide a large annotated dataset as well as an organized platform to compare algorithmic methods in the use case of the segmentation of enlarged lymph nodes in the mediastinum. This is specifically relevant for lung cancer patients, but can certainly serve as a benchmark for the extension to other lymph nodes in the human body. The mediastinum contains ten or more lymph nodes, the positioning of which is defined in El-Sherief et al. (2014).
The problem setting for this challenge consists of three main hurdles that need to be addressed by the participant’s approaches: First, the provided data in combination with other publicly available datasets is highly heterogeneous regarding the retrieval process and image characteristics such as variances in image resolution and field of view. In addition, the provided challenge data shows images of patients with pathologies (e.g. a collapsed lung, tumors) in the highly individual anatomy of the mediastinum, which makes automatic segmentation and registration particularly difficult. Second, finding lymph nodes is a classic “looking for a needle in a haystack” problem, as lymph nodes are small. Therefore, the amount of foreground pixels is also small compared to the number of background pixels, which leads to a severe class imbalance. Third, the training dataset for the challenge is weakly annotated, providing segmentation masks of only one or more clinically relevant lymph nodes. The overall results of the challenge show that not accounting for the problem of undersegmentation will result in a significant performance drop on validation and test set.
This study centers around the use of multiple spatial anatomical priors integrated as supplemental inputs into deep learning methodologies as a tool to address challenges mentioned above. The priors consist of distance maps normalized to the atlas’ coordinate system and probability maps indicating the likelihood of lymph node occurrence. To generate the priors, we developed an upstream atlas-to-patient registration approach. The extensive use of additional image augmentation improves generalizability, bridges the domain gap between different datasets, and, in this way, tackles the high data heterogeneity. To address the problem of a high false negative rate due to the strong class imbalance and partial annotation, we use the probabilistic lymph node atlas in loss weighting and post-processing.
2 Related Works
The field of automatic lymph node classification, detection, and segmentation has a rich history in medical research. Feuerstein et al. (2012) advanced the generation of an atlas, originally designed for brain imaging, for lymph nodes in the mediastinum to then detect and label the lymph node stations. Similarly, Feulner et al. (2013) developed a probabilistic atlas derived from lymph node segmentation masks, employing it as a spatial prior in a multistep approach based on conventional methods. To refine the prior, the authors smooth the resulting average lymph node segmentations and exclude selected organs, i.e. lungs, the trachea, the esophagus, and the heart.
The landscape shifted with Roth et al. (2014), who contributed a dataset of 3D CT volumes from 90 patients with 388 segmented lymph nodes, propelling the rise of learning-based methods in this domain. Roth et al. (2014) trained a convolutional neural network using a 2.5D resampling strategy. As input, the authors use three randomly scaled, rotated and translated orthogonal 2D slices centered on a volume of interest’s centroid coordinates. Subsequent researchers, such as Iuga et al. (2021) and Nayan et al. (2022), expanded on this work by experimenting with diverse network architectures. Iuga et al. (2021) introduced a 3D fully convolutional foveal neural network capable of extracting features at various resolutions, while Nayan et al. (2022) explored a modified upsampling strategy for U-Net++. Oda et al. (2018) trained a 3D U-Net to segment lymph nodes as well as other anatomical structures to prevent oversegmentation.
A promising integration of spatial prior information and learning-based approaches was proposed by Bouget et al. (2021). The authors incorporated segmentation masks of anatomical structures, such as the esophagus and the azygos vein, as additional channel input to a 3D U-Net. This strategic inclusion aims to prevent the network from generating false positives in these specific areas. In this way, the authors synthesize the strengths of both worlds – deep learning methods and anatomical priors.
3 Methods
Our approach consists of a pre-processing step, the network training and a post-processing of the predicted results. In the pre-processing, automatic segmentation of anatomical structures is followed by atlas-to-patient registration to generate strong anatomical priors, which are used as additional network input, in loss weighting, and in the post-processing step. A U-Net architecture was trained as a segmentation model. The overall framework is shown in Fig. 1.
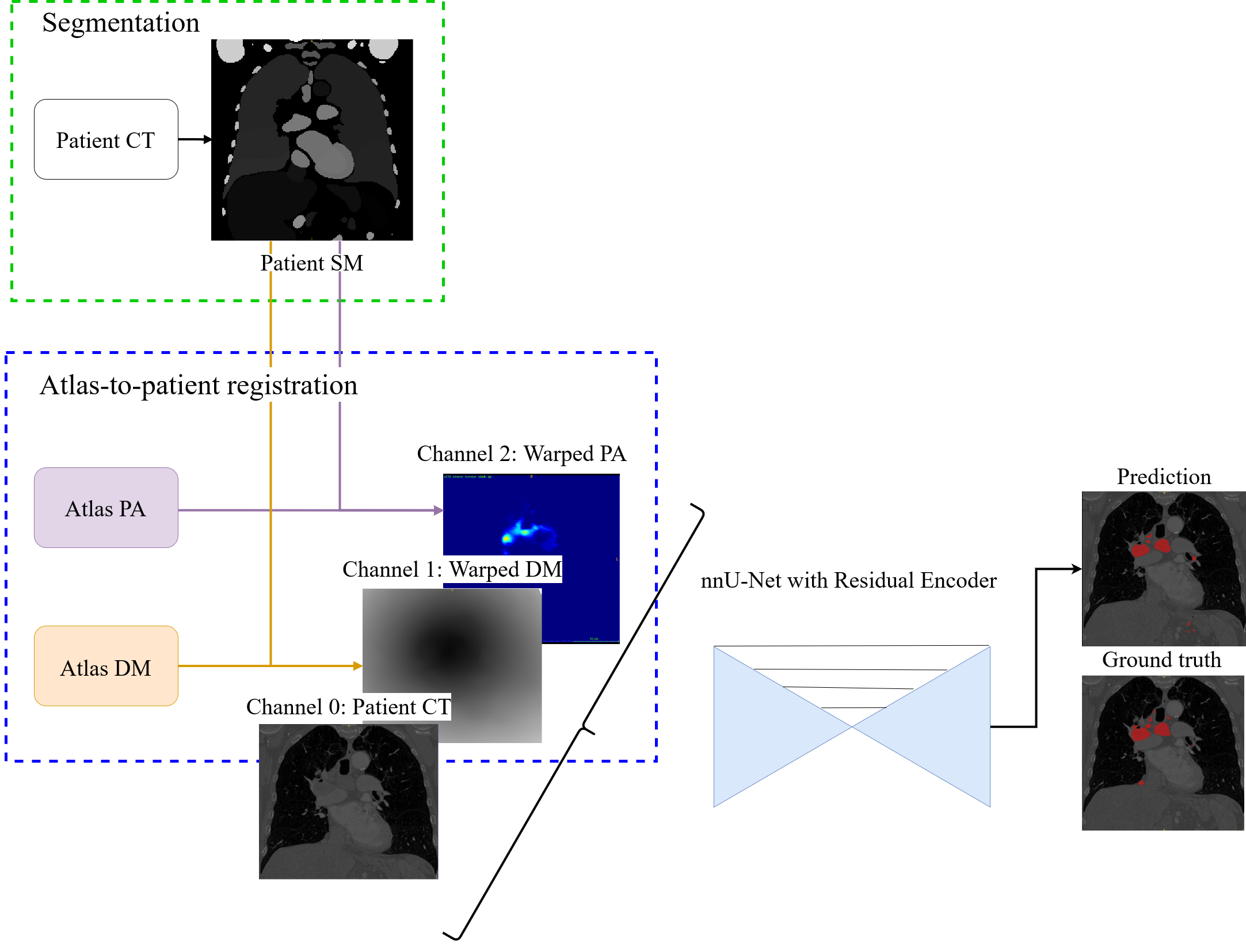 |
3.1 Pre-Processing and Generation of Anatomical Priors
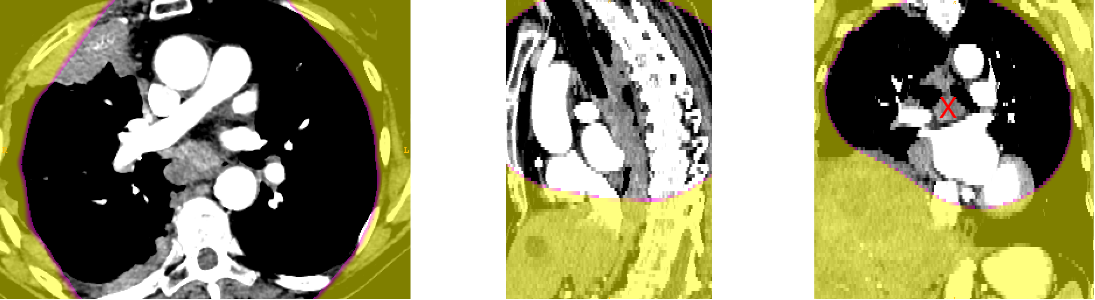
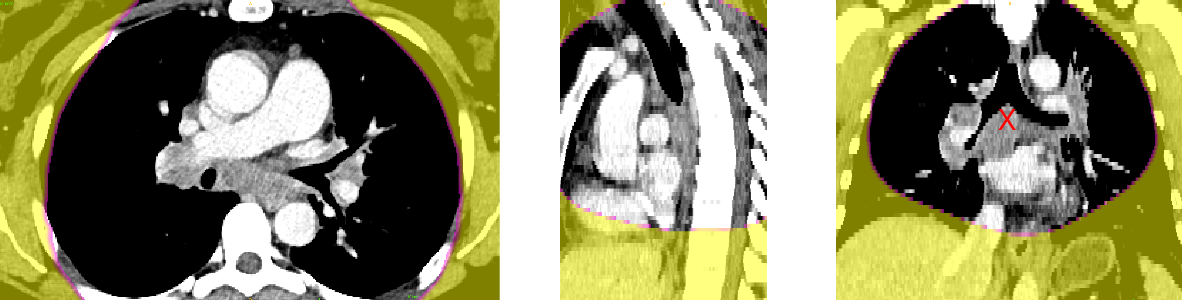
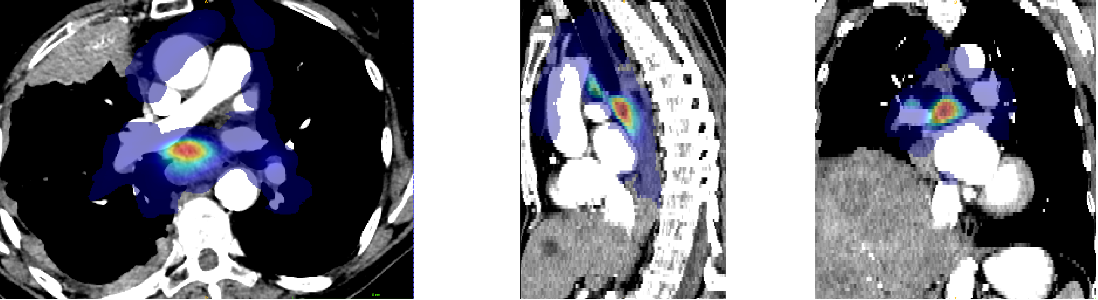
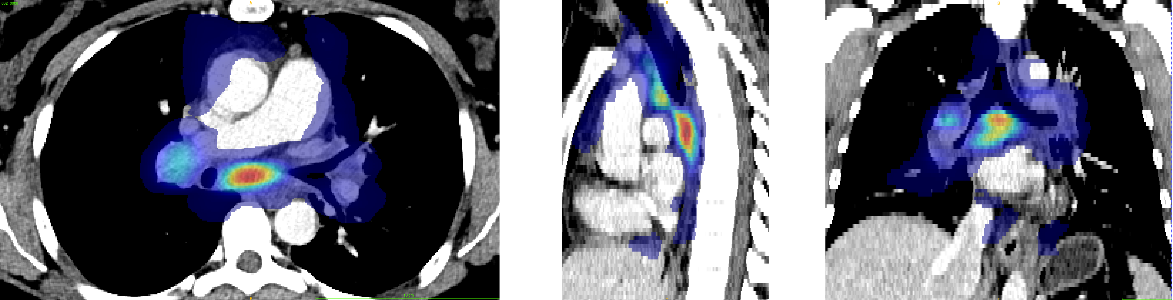
Anatomical prior information was introduced to account for the low contrast of lymph nodes in CT images, strong class imbalance and incomplete labelling of the training data. The proposed prior information consists of anatomical labelling, a distance map (DM) calculated with respect to a specified anatomical landmark (bifurcation of the trachea) defining a kind of anatomical coordinate system, and a probability map for the occurrence of malignant lymph nodes in the training set, referred to as probabilistic lymph node atlas (PA). Visualizations for both priors for example patients are shown on Fig. 2 and Fig. 3.
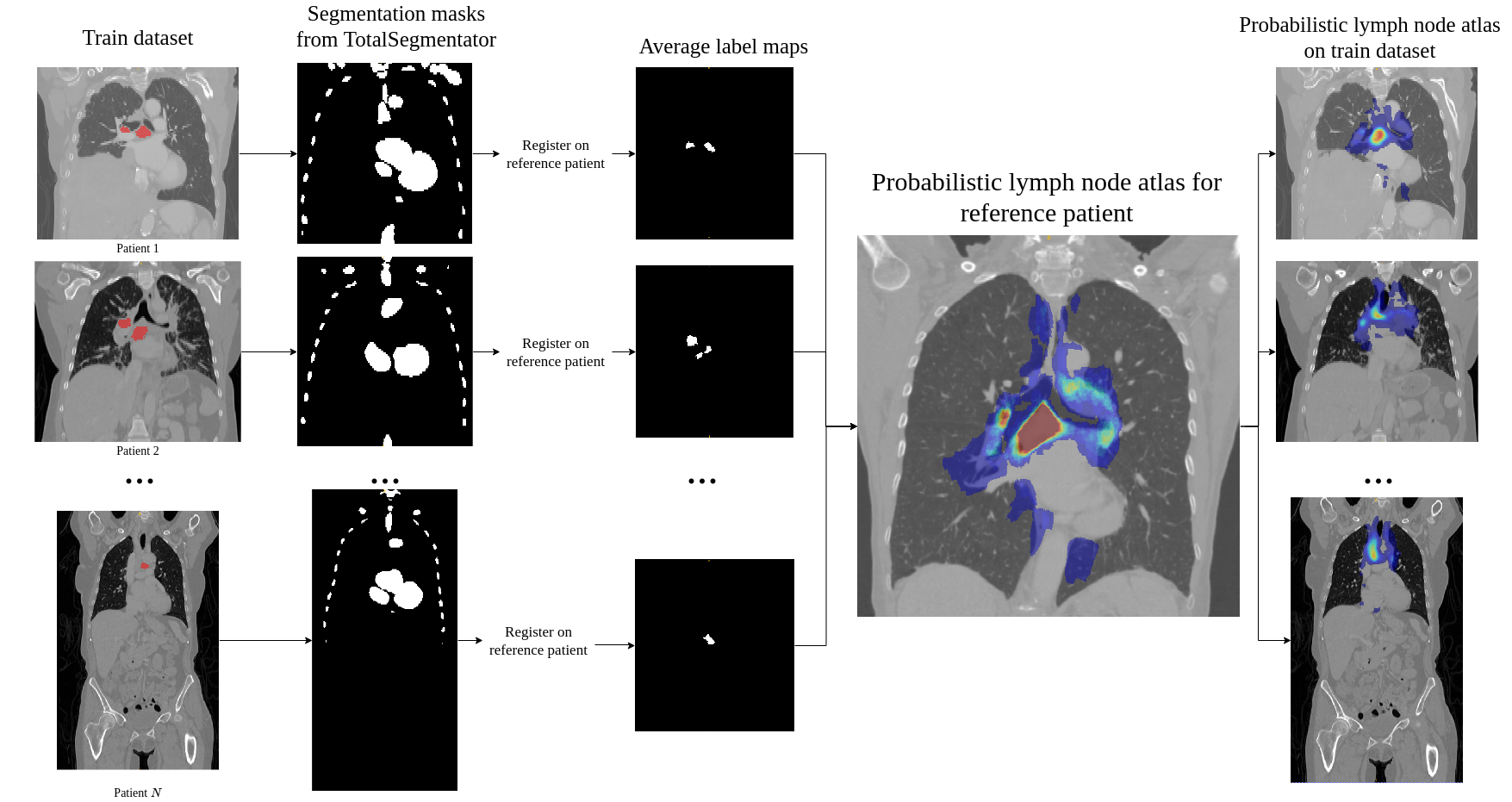
To generate the probabilistic lymph node atlas, all training images were registered to a chest CT lymph node atlas (Lynch et al., 2013) and their ground truth annotations were used to compute occurrence probabilities. The registration pipeline consisted of a sequence of a rigid registration, followed by an affine registration, and finally a deformable registration using a GPU-based implementation of ITK’s VariationalRegistration111https://itk.org/Doxygen/html/group__VariationalRegistration.html module (Werner et al., 2014). The strong heterogeneity of the CT data, as mentioned in Sec. 1, would compromise the robustness of pure intensity-based registration. Therefore, rigid and affine registration were based on segmentation masks (SM) of selected anatomical structures segmented by the TotalSegmentator222https://github.com/wasserth/totalsegmentator algorithm (Wasserthal et al., 2023). The selected structures (i.e. bones, heart, esophagus, trachea, and aorta) include anatomies that are less likely to contain pathologies, which could distort registration results. The resulting occurrence probabilities were smoothed with a Gaussian filter () and then scaled to a range between zero and one. The distance map was also defined in the atlas space with respect to the selected reference point and normalized to a range between zero and one.
For training and inference, the chest CT atlas was registered to the input images using the registration pipeline described above, and the resulting deformation was used to transfer the probabilistic lymph node atlas and distance map to the subject’s coordinate system. Further, the image data was cropped to the bounding box of the lung segmentation masks to reduce computational costs and intensity normalization was performed using nnU-Net’s default CT normalization method, which involved taking the 0.5 and 99.5 percentiles of intensity values within the foreground class. This yields values resembling the soft tissue intensity window.
3.2 Model Architecture and Loss Calculation
As a basis for our segmentation network, we used nnU-Net by Isensee et al. (2021). Inspired by Isensee et al. (2023), we included additional residual connections in the encoder of the U-Net. The loss function was a combination of Dice loss, Cross-Entropy (CE) loss and Tversky loss (Salehi et al., 2017):
| (1) |
where is the image space, and and the ground truth and predicted label maps. The loss weights were chosen to be , , and . The Tversky loss addresses the class imbalance of fore- and background. By adjusting its two hyperparameters, that is, setting and , we allow a higher false positive rate.
Additionally, we introduced a loss weighting according to the probabilistic lymph node atlas to further account for the partial annotation of the training data. Due to the partial annotation, false positive predictions can actually be true positives, which are missing in the ground truth annotation. This approach uses a reduced Dice loss weight in areas with high lymph node occurrence. The weight map at pixel index was calculated as follows:
| (2) |
where is the dilated ground truth segmentation with a kernel radius of and is the probabilistic lymph node atlas. The ground truth segmentations were dilated to ensure that the lymph node borders are correctly classified. The resulting weight map was multiplied with the pixel-wise Dice loss. This was carried out for multiple resolutions, as loss calculation of the nnU-Net is embedded into a deep supervision framework.
3.3 Augmentation
nnU-Net uses standard augmentations such as random cropping, rotating, and flipping per default. To further enable training with different CT data sets, we adapted the single-source domain generalization technique from Ouyang et al. (2021). This allows bridging the domain gap caused by different acquisition processes and scanners. During each training iteration, two random intensity or texture transformations were sampled from a shallow convolutional network to generate a global intensity non-linear augmentation (GIN) for each input image. Both GIN augmentations were then merged using interventional pseudo-correlation augmentation (IPA). More precisely, a bias-field-like pseudo-correlation map was used to perform the blending. See the work of Ouyang et al. (2021) for more details. The impact of the GIN and IPA augmentation was controlled by a blending parameter whose weighting follows a Gaussian ramp-up curve (Laine and Aila, 2017) for the first 1,000 epochs of training to gradually introduce domain generalization to the nnU-Net.
3.4 Post-processing
The aim of the proposed post-processing steps is to, on the one hand, ensure that only enlarged lymph nodes are segmented and, on the other hand, account for the partial annotations and strong class imbalance. Consequently, the post-processing consisted of removing connected components that have a diameter smaller than 3, 5 or 7 mm or skipping this post-processing step. Additionally, we set the threshold for binarization for pixel according to:
| (3) |
where is the threshold depending on the probabilistic lymph node atlas at pixel , is the constant threshold, and is a pixel at index of the probabilistic lymph node atlas . Similarly to Bouget et al. (2021), the threshold was kept at or reduced to or . This allows for more uncertainty in areas where lymph nodes are more likely to occur and results in enlarged segmentation masks. In addition, it was necessary to revert the cropping to the lung segmentation mask by padding the predicted masks to the original input size and removing segmented pixels outside the convex hull of the lung.
Furthermore, we utilized random variations in network training by ensembling the predictions of five models to account for partial annotation and class imbalance.
3.5 Semi-Supervised Learning
The presented approach was trained in a purely supervised manner. However, there is potential to improve generalization by using unsupervised learning techniques. To examine this, we tested several modifications of the semi-supervised learning approach introduced by Wang et al. (2022). The approach comes from the field of computer vision, and the transferability to the medical image domain has not been researched before. The main idea of the original approach can be summarized as follows: Given two networks with identical network architectures – a student and a teacher, the student is trained given labeled data as well as unlabeled data with pseudo-labels generated by the teacher. The teacher weights are updated by computing an exponential moving average of the weights of the student. The teacher’s task is to differentiate between reliable and unreliable segmented pixels by computing the entropy of the softmax probabilities of class (Wang et al., 2022). A pixel is reliable, if exceeds a dynamically increasing threshold, which is computed based on the histogram of entropies. In this way, the teacher generates pseudo-labels for unlabeled data to diversify the training data for the student’s learning process.
The first modification we implemented is a strong augmentation for the student, including GIN and IPA, as well as additional random contrast adjustment and random Gaussian noise. The unlabeled data that was fed to the teacher was not augmented, besides the standard augmentations of nnU-Net. This should ensure the best possible pseudo-label generation by the teacher and a robust segmentation performance through learning on diverse training data by the student.
The downside of the original approach in combination with partially annotated data and the strong class imbalance is that the foreground pixels at the borders of the lymph nodes are often considered to be unreliable. The reason is that the network is more uncertain in detecting lymph nodes than in detecting background. This leads to smaller pseudo-labels, which in turn result in degrading performance. Hence, we developed a second variant that accounts for the problem of undersegmentation. We removed the differentiation between reliable and unreliable pixels and instead use our proposed post-processing approach described in Sec. 3.4 for pseudo-label generation. This strategy artificially enlarges the segmentation masks predicted by the teacher, which in the process of training should lead to the predicted lymph node segmentations to become bigger instead of smaller.
4 Results
The chosen nnU-Net configuration was to train on patches of size 128112160 with a batch size of two. The learning rate was set to linearly decrease from 0.01 to until epoch 1,000 and from to until epoch 2,000. This scheduling leads to a more rapid decrease of the learning rate in the first training half than in the second training half. To increase computational efficiency, we implemented a smart cache strategy. More precisely, the items used for training are stored in cache and partially replaced in each iteration.
The results for supervised training are described in the following sections. For the training of the semi-supervised learning task, we resumed training from the best checkpoint of one of the pre-trained folds. Even though the implementation of the semi-supervised approaches was particularly time-consuming, all proposed implementations led to a degradation of the validation accuracy during training. Typical learning curves can be seen in Fig. 4.
4.1 Data
We used three different datasets for network training: First, 393 partially annotated CT images from the Mass General Brigham Hospital in Boston are provided as the challenge training dataset (Khajavibajestani et al., 2023). Second, a total of 89 patients are part of a dataset collected by the National Institutes of Health in Maryland (Roth et al., 2014). Even though, this dataset contains annotations, they have been refined by Bouget et al. (2021). The annotations of Bouget et al. (2021) for this dataset as well as another 30 patients from St. Olavs hospital in Trondheim (third dataset) contain lymph nodes of different sizes as well as information about the node’s station. For a non-expert, it is difficult to distinguish the segmentation of stations opposed to the segmentation of single lymph nodes with identifiable borders. The inability to properly detect instances and separate collocated lymph nodes as a limitation was addressed and analyzed in Bouget et al. (2021). The image resolution of the training data varies between and in-plane and to between slices. The field of view can either be limited to the patient’s thorax, but can also extend to the whole body. If not marked otherwise, the trained models were tested on the test dataset of the LNQ 2023 challenge. The test set contains 100 patients and a total of 861 segmented lymph nodes in the ground truth annotations. Some additional characteristics of the described datasets can be reviewed in Tab. 1. The lymph node size is the minimum diameter of an ellipse fit to a fully connected component, that is, the segmentation mask of a single lymph node. The minimum ellipsoid diameter can differ slightly from the in-plane diameter as defined in RECIST.
Dataset# PatientsTotal # LNAVG # LNAVG LN sizeRoth et al. (2014)89135115.188.5482Bouget et al. (2021)3052617.536.6453LNQ train3935561.4116.5481LNQ val201055.259.2483LNQ test1008618.618.1831
4.2 Metrics
The evaluation metrics, Dice and Average Symmetric Surface Distance (ASSD), were chosen by the challenge organizers. Tab. 2 shows the results of our final submission for the LNQ 2023 challenge for validation and test set. Here, we used an ensemble of five models, three of which were trained with the PA-weighted Dice loss and two were trained without. The threshold for binarization was set to and the minimum diameter of the detected lymph nodes was 5 mm.

| Dataset | Dice | ASSD |
|---|---|---|
| LNQ val | 0.5600 | 6.7905 |
| LNQ test | 0.5690 | 6.8976 |
Additionally, we report Precision and Recall, as well as the percentage of lymph nodes that were correctly found. The metric LN found shows the fraction of predicted lymph nodes overlapping with the segmented ground truth lymph nodes, i.e. . The calculation of this metric was based on the implementation introduced by the organizers of the BRATS 2023 challenge, initially used for the evaluation of lesion segmentation in brain images.333https://github.com/rachitsaluja/BraTS-2023-Metrics To assess the overlap of the predicted and the ground truth masks, a connected component analysis is carried out based on dilated masks. The dilation factor was set to two.
4.3 Ablation Study
In the following, we present an ablation study to quantify the effects of the proposed modifications to the baseline model, which is using the standard nnU-Net trainer with minor modifications to the learning rate and the epoch number on the provided CT data. Models were trained and averaged over five model runs. Training data, network architecture, and training strategy were kept the same for each process.
The results with and without post-processing are presented in Tab. 3 and Tab. 4 respectively. The hyperparameters for post-processing were set via grid search. Lowering the threshold for binarization to without any restriction of the minimum lymph node diameter consistently improves the Dice score and Recall. For significance testing, a one-sided t-test (alternative hypothesis: the mean of the distribution underlying the first sample is greater/lower than the mean of the distribution underlying the second sample) has been carried out. The two prior options, that is the additional segmentation masks or the distance map in combination with the probabilistic lymph node atlas, were tested against using only CT as input for training. All other models were compared to the preceding model.
The use of both prior options leads to an improved Dice score. However, only using SM as a prior yields a p-value smaller than in the one-sided t-test while using DM & PA as priors is not significant. Surprisingly, a study based on solely fully annotated data showed the opposite effect for using SM as a prior. In this study, the Dice score is approximately three percentage points lower than training on CT only (Engelson et al., 2024). All other suggested configurations improve segmentation performance significantly. A large effect is achieved by accounting for the partial annotation by oversampling the fully annotated datasets, as well as using the Tversky loss to reduce the false negative rate. Tackling the high data heterogeneity by using GIN and IPA as an additional augmentation technique strongly improves performance. The last suggested configuration is using the PA-weighted Dice loss calculation, which, as expected, reduces the amount of false negatives. Generally, the suggested training modifications and post-processing lead to a decreasing false negative rate at the price of an increasing false positives rate. In other words, the Recall improves continuously while maintaining a stable or small decrease in Precision. When training on CT only, around 27% of the ground truth lymph nodes are found by the model. The combination of all suggested approaches enables the algorithm to find 57% of the ground truth lymph nodes.
DMPASMOvers.& Tver. LossGIN &IPAPA-weigh.lossDiceASSDPrecisionRecallLN found0.3531 0.007714.6567 0.94420.81940.25630.2652X0.3818 0.012712.8515 0 5,9950.81070.27700.2732XX0.3611 0.013814.1947 0.85830.83630.26840.2656XXX0.4297 0.003411.7228 0.57040.83820.30620.3104XXXX0.5269 0.04287.6773 1.73140.80610.42830.4448XXXXX0.5509 0.00536.9804 0 31090.79680.45900.4763ensemble0.55546.82490.81480.45980.4586
DMPASMOvers.& Tver. LossGIN &IPAPA-weigh.lossDiceASSDPrecisionRecallLN found0.3758 0.007114.3196 0.98790.79830.28150.2749X0.4024 0.012213.0008 1.07900.78810.30200.2807XX0.3825 0.012814.4255 1.09310.81510.29410.2754XXX0.4524 0.002911.0104 .0.61280.82280.33440.3299XXXX0.5491 0.04237.1048 1.66490.78530.46430.4896XXXXX0.5703 0.0066.4780 0.33970.77380.49430.5153ensemble0.60335.74830.76060.54830.5654
With a Dice score of the submitted model (Tab. 2) has a lower performance than the best model with a Dice score of displayed in Tab. 4. This portrays the following two problems when developing algorithms embedded in a challenge setting: First, the hyperparameter search was not performed systematically, as participants had no access to the fully annotated challenge datasets. Each submission required effort and the number of submissions was limited. And second, hyperparameters for post-processing optimized on the small validation set did not generalize to the test set. As can be reviewed in Tab. 1, the characteristics such as average lymph nodes size differ in train, validation, and test set.
5 Discussion
For the task of mediastinal lymph node segmentation, we propose a modified nn-UNet with the use of anatomical priors as additional model input, in loss weighting, and post-processing. The priors are generated with an atlas-to-patient registration approach and serve as orientation reference (distance map) as well as feature guidance (probabilistic lymph node atlas). The ablation study in Sec. 4.3 shows that using anatomical priors provided as additional input alone does not lead to a significant performance gain. However, the probabilistic lymph node atlas enhances loss calculation and post-processing, as well as successfully addresses the challenge of partial annotation and class imbalance. The use of additional (fully annotated) training data and augmentation techniques show the largest effects on the segmentation accuracy. However, using the probabilistic lymph node atlas for post-processing and loss weighting reduces the number of false negatives, which is oftentimes relevant for medical tasks and when only weakly annotated training data is available.
In the following section, we discuss possible reasons for why the use of the publicly available fully annotated data turned out to be crucially important to yield decent segmentation accuracy. This could also serve as justification for the failed semi-supervised learning approach for the application of mediastinal lymph node segmentation.
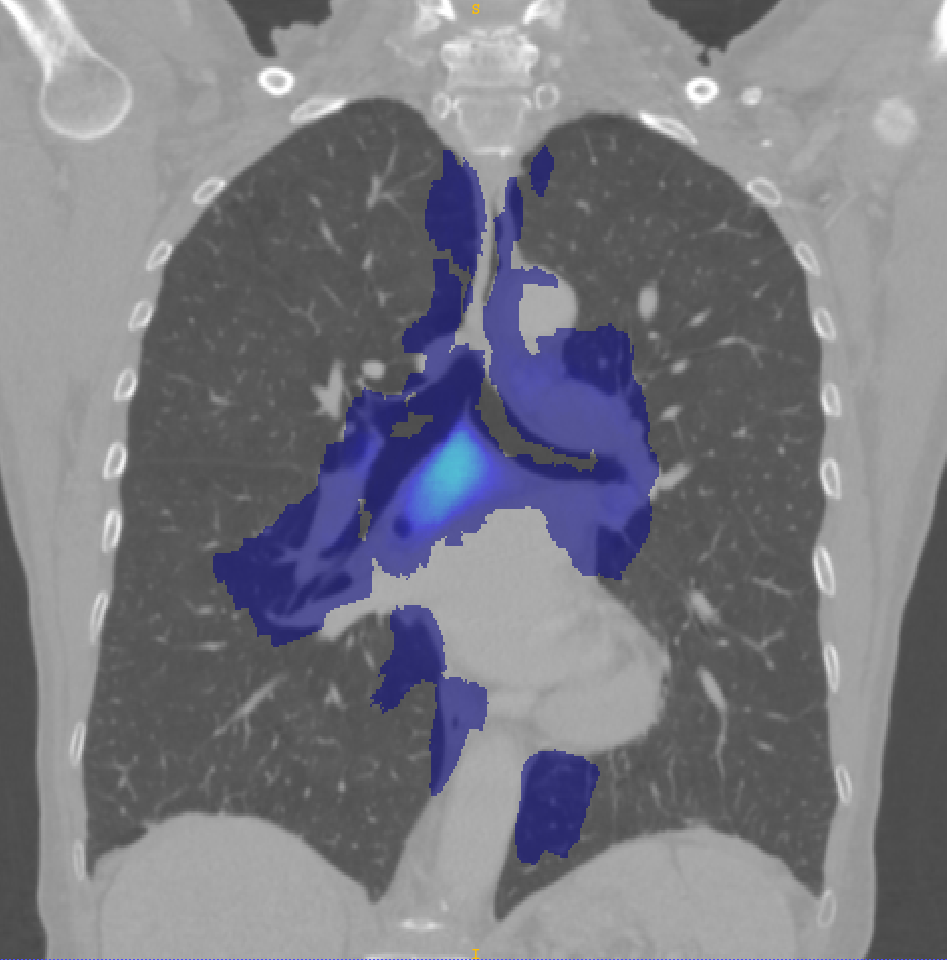

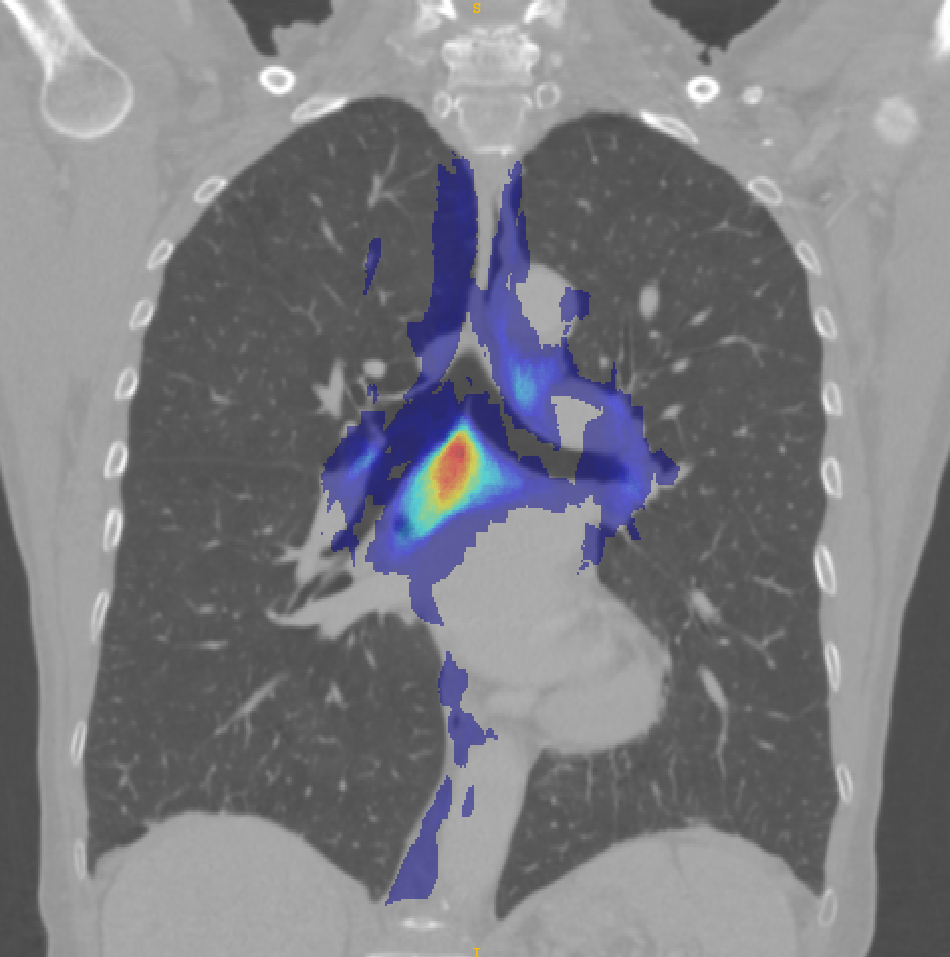

Figure 5 shows the probabilistic lymph node atlases in coronal view generated with the segmentation masks of the LNQ 2023 train dataset in 5(a), the validation dataset in 5(b), and the test dataset in 5(c). According to these images, we find the following differences between train, validation and test dataset:
- •
The test set includes lymph node stations that are not included in the train set, e.g. between the bottom right lung and the aorta.
- •
Not all stations occur equally often in train, validation and test, e.g. station 7 (beneath the bifurcation of the trachea) and 4L (between aorta and trachea) is segmented more often in val/test than in train.
- •
The size of the segmented masks differs quite heavily, e.g. the segmentation masks in the train set are larger on average. This is confirmed and analyzed in more detail by a co-participant (Fischer et al., 2024).
It can be concluded that the data distribution of the partially annotated train set differs from the fully annotated validation and test set because the choice of which lymph nodes to partially annotate has not been made randomly. On the one hand, it can be expected that medical experts are more likely to annotate strongly enlarged lymph nodes instead of lymph nodes that are pathological, but smaller. On the other hand, some stations are easier to find in the mediastinum than others and, therefore, are examined more often, e.g. the location of station 7 is distinct for most patients and pathologies. Self-enhancing methods, such as the proposed semi-supervised learning approach, reproduce results learned the by the model trained in a supervised manner on the data distribution of the training data. If the data or annotations of the training dataset carry biases, they get enforced with further unsupervised training. More extensive hyperparameter tuning could have possibly improved results by tackling the problem of a high false negative rate, or in other words ”the problem of not segmenting enough”. However, this, most probably, would have not mitigated the consequences of the biased partial annotation, i.e. ”the problem of not segmenting the right thing”.
The incorporation of anatomical priors, or in some way encoded prior knowledge, into the learning process of deep learning methods does seem intuitive when dealing with tasks of high complexity and label uncertainty, such as in the task presented in the LNQ 2023 challenge. It is necessary to point out that the anatomical priors were only provided as additional inputs to the algorithm, without controlling whether the algorithm learns features from them. Possibly, CT already contains all necessary information to segment pathological lymph nodes. Simultaneously, the segmentation performance does have potential for improvement – the best performing model still misses to segment over 40% of the pathological lymph nodes. In medical image applications, oftentimes, training datasets are small, but the task complexity is high. Therefore, algorithms are particularly at risk of showing the ”Clever Hans” behavior, that is predictors making correct predictions for the wrong reasons (Lapuschkin et al., 2019). Examples of this are shown in DeGrave et al. (2021), Badgeley et al. (2019), and Zech et al. (2018). Also, in the application of mediastinal lymph node segmentation, it is difficult to say, whether the features encoded in the U-Net encoder are semantically meaningful. The symbiosis of prior knowledge with the learning process has the potential to ensure that the learned features actually are responsible for class changes. Therefore, it is necessary to explore other options of incorporating prior knowledge in the network learning process in further research. To further improve segmentation accuracy for the underlying use case, it might be interesting to use the fully annotated validation and test data for training.
Acknowledgments
This work was supported by grants of the collaborative research project “AI ecosystem in health care” within the subproject titled “Health system-based analysis of disease patterns using lung diseases as an example” financed by the federate state Schleswig-Holstein, Germany.
Ethical Standards
The work follows appropriate ethical standards in conducting research and writing the manuscript, following all applicable laws and regulations regarding treatment of animals or human subjects.
Conflicts of Interest
We declare we don’t have conflicts of interest.
References
- Badgeley et al. (2019) Marcus A. Badgeley, John R. Zech, Luke Oakden-Rayner, Benjamin S. Glicksberg, Manway Liu, William Gale, Michael V. McConnell, Bethany Percha, Thomas M. Snyder, and Joel T. Dudley. Deep learning predicts hip fracture using confounding patient and healthcare variables. npj Digital Medicine, 2(1):1–10, 2019. ISSN 2398-6352.
- Bouget et al. (2021) David Bouget, André Pedersen, Johanna Vanel, Håkon Olav Leira, and Thomas Langø. Mediastinal lymph nodes segmentation using 3D convolutional neural network ensembles and anatomical priors guiding. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 11:44 – 58, 2021.
- DeGrave et al. (2021) Alex J. DeGrave, Joseph D. Janizek, and Su-In Lee. AI for radiographic COVID-19 detection selects shortcuts over signal. Nature Machine Intelligence, 3(7):610–619, 2021. ISSN 2522-5839.
- Eisenhauer et al. (2009) E.A. Eisenhauer, P. Therasse, Bogaerts J., L.H. Schwartz, D. Sargent, R. Ford, J. Dancey, S. Arbuck, S. Gwyther, M. Mooney, L. Rubinstein, L. Shankar, L. Dodd, R. Kaplan, D. Lacombe, and J. Verweij. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer, 45(2):228–247, 2009. ISSN 0959-8049. Response assessment in solid tumours (RECIST): Version 1.1 and supporting papers.
- El-Sherief et al. (2014) Ahmed H. El-Sherief, Charles T. Lau, Carol C. Wu, Richard L. Drake, Gerald F. Abbott, and Thomas W. Rice. International Association for the Study of Lung Cancer (IASLC) Lymph Node Map: Radiologic Review with CT Illustration. RadioGraphics, 34(6):1680–1691, 2014. PMID: 25310423.
- Engelson et al. (2024) Sofija Engelson, Jan Ehrhardt, Joshua Niemeijer, Stefanie Schierholz, Lennart Berkel, Malte Maria Elser, Yannic Sieren, and Heinz Handels. Comparison of anatomical priors for learning-based neural network guidance for mediastinal lymph node segmentation. In Weijie Chen and Susan M. Astley, editors, Medical Imaging 2024: Computer-Aided Diagnosis, volume 12927, page 1292719. International Society for Optics and Photonics, SPIE, 2024.
- Feuerstein et al. (2012) Marco Feuerstein, Ben Glocker, Takayuki Kitasaka, Yoshihiko Nakamura, Shingo Iwano, and Kensaku Mori. Mediastinal atlas creation from 3-D chest computed tomography images: Application to automated detection and station mapping of lymph nodes. Medical Image Analysis, 16(1):63–74, 2012. ISSN 1361-8415.
- Feulner et al. (2013) Johannes Feulner, S. Kevin Zhou, Matthias Hammon, Joachim Hornegger, and Dorin Comaniciu. Lymph node detection and segmentation in chest CT data using discriminative learning and a spatial prior. Medical Image Analysis, 17(2):254–270, 2013. ISSN 1361-8415.
- Fischer et al. (2024) Stefan M. Fischer, Johannes Kiechle, Daniel M. Lang, Jan C. Peeken, and Julia A. Schnabel. Mask the Unknown: Assessing Different Strategies to Handle Weak Annotations in the MICCAI2023 Mediastinal Lymph Node Quantification Challenge. Machine Learning for Biomedical Imaging, 2:798–816, 2024. ISSN 2766-905X.
- Isensee et al. (2021) Fabian Isensee, Paul F Jaeger, Simon A A Kohl, Jens Petersen, and Klaus H Maier-Hein. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2):203—211, 2021. ISSN 1548-7091.
- Isensee et al. (2023) Fabian Isensee, Constantin Ulrich, Tassilo Wald, and Klaus H. Maier-Hein. Extending nnU-Net Is All You Need. In Thomas M. Deserno, Heinz Handels, Andreas Maier, Klaus Maier-Hein, Christoph Palm, and Thomas Tolxdorff, editors, Bildverarbeitung für die Medizin 2023, pages 12–17, Wiesbaden, 2023. Springer Fachmedien Wiesbaden. ISBN 978-3-658-41657-7.
- Iuga et al. (2021) Andra Iuga, Heike Carolus, Anna Höink, Tom Brosch, Tobias Klinder, David Maintz, Thorsten Persigehl, Bettina Baeßler, and Michael Püsken. Automated detection and segmentation of thoracic lymph nodes from CT using 3D foveal fully convolutional neural networks. BMC Medical Imaging, 21, 04 2021.
- Khajavibajestani et al. (2023) Roya Khajavibajestani, Steve Pieper, Erik Ziegler, Tagwa Idris, Reuben Dorent, Bhanusupriya Somarouthu, Sonia Pujol, Ann LaCasce, Heather Jacene, Gordon Harris, and Ron Kikinis. Mediastinal Lymph Node Quantification (LNQ): Segmentation of Heterogeneous CT Data, 2023.
- Laine and Aila (2017) Samuli Laine and Timo Aila. Temporal Ensembling for Semi-Supervised Learning. In 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, Conference Track Proceedings. OpenReview.net, 2017.
- Lapuschkin et al. (2019) Sebastian Lapuschkin, Stephan Wäldchen, Alexander Binder, Grégoire Montavon, Wojciech Samek, and Klaus-Robert Müller. Unmasking clever hans predictors and assessing what machines really learn. Nature Communications, 10, 03 2019.
- Lynch et al. (2013) Rod Lynch, Graham Pitson, David Ball, Line Claude, and David Sarrut. Computed tomographic atlas for the new international lymph node map for lung cancer: A radiation oncologist perspective. Practical Radiation Oncology, 3(1):54–66, January 2013. ISSN 18798500.
- Nayan et al. (2022) Al-Akhir Nayan, Boonserm Kijsirikul, and Yuji Iwahori. Mediastinal Lymph Node Detection and Segmentation Using Deep Learning. IEEE Access, 10:89289–89307, 2022.
- Oda et al. (2018) Hirohisa Oda, Holger R. Roth, Kanwal K. Bhatia, Masahiro Oda, Takayuki Kitasaka, Shingo Iwano, Hirotoshi Homma, Hirotsugu Takabatake, Masaki Mori, Hiroshi Natori, Julia A. Schnabel, and Kensaku Mori. Dense volumetric detection and segmentation of mediastinal lymph nodes in chest CT images. In Nicholas Petrick and Kensaku Mori, editors, Medical Imaging 2018: Computer-Aided Diagnosis, volume 10575, page 1057502. International Society for Optics and Photonics, SPIE, 2018.
- Ouyang et al. (2021) Cheng Ouyang, Chen Chen, Surui Li, Zeju Li, Chen Qin, Wenjia Bai, and Daniel Rueckert. Causality-Inspired Single-Source Domain Generalization for Medical Image Segmentation. IEEE Transactions on Medical Imaging, 42:1095–1106, 2021.
- Roth et al. (2014) Holger R. Roth, Le Lu, Ari Seff, Kevin M. Cherry, Joanne Hoffman, Shijun Wang, Jiamin Liu, Evrim Turkbey, and Ronald M. Summers. A New 2.5D Representation for Lymph Node Detection Using Random Sets of Deep Convolutional Neural Network Observations. In MICCAI 2014, pages 520–527. Springer, 2014. ISBN 978-3-319-10404-1.
- Salehi et al. (2017) Seyed Sadegh Mohseni Salehi, Deniz Erdogmus, and Ali Gholipour. Tversky Loss Function for Image Segmentation Using 3D Fully Convolutional Deep Networks. In Qian Wang, Yinghuan Shi, Heung-Il Suk, and Kenji Suzuki, editors, Machine Learning in Medical Imaging, pages 379–387, Cham, 2017. Springer International Publishing. ISBN 978-3-319-67389-9.
- Wang et al. (2022) Yuchao Wang, Haochen Wang, Yujun Shen, Jingjing Fei, Wei Li, Guoqiang Jin, Liwei Wu, Rui Zhao, and Xinyi Le. Semi-supervised semantic segmentation using unreliable pseudo-labels. In 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pages 4238–4247, 2022.
- Wasserthal et al. (2023) Jakob Wasserthal, Hanns-Christian Breit, Manfred T. Meyer, Maurice Pradella, Daniel Hinck, Alexander W. Sauter, Tobias Heye, Daniel Boll, Joshy Cyriac, Shan Yang, Michael Bach, and Martin Segeroth. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. Radiology: Artificial Intelligence, 2023.
- Werner et al. (2014) René Werner, Alexander Schmidt-Richberg, Heinz Handels, and Jan Ehrhardt. Estimation of lung motion fields in 4D CT data by variational non-linear intensity-based registration: A comparison and evaluation study. Physics in Medicine & Biology, 59(15):4247, July 2014.
- Zech et al. (2018) John R. Zech, Marcus A. Badgeley, Manway Liu, Anthony B. Costa, Joseph J. Titano, and Eric Karl Oermann. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Medicine, 15(11):e1002683, 2018. ISSN 1549-1676.
A Generation of the Probabilistic Lymph Node Atlas
 |

