Echocardiography Analysis with Deep Learning using Priors: Multi-centric Evaluation of Generalisation
Yingyu Yang1 , Marie Rocher2, Pamela Moceri2
, Marie Rocher2, Pamela Moceri2 , Maxime Sermesant1
, Maxime Sermesant1
1: Centre Inria d’Université Côte d’Azur, Sophia Antipolis, France, 2: CHU de Nice - Hôpital Pasteur, Nice, France
Publication date: 2024/11/30
https://doi.org/10.59275/j.melba.2024-58fd
Abstract
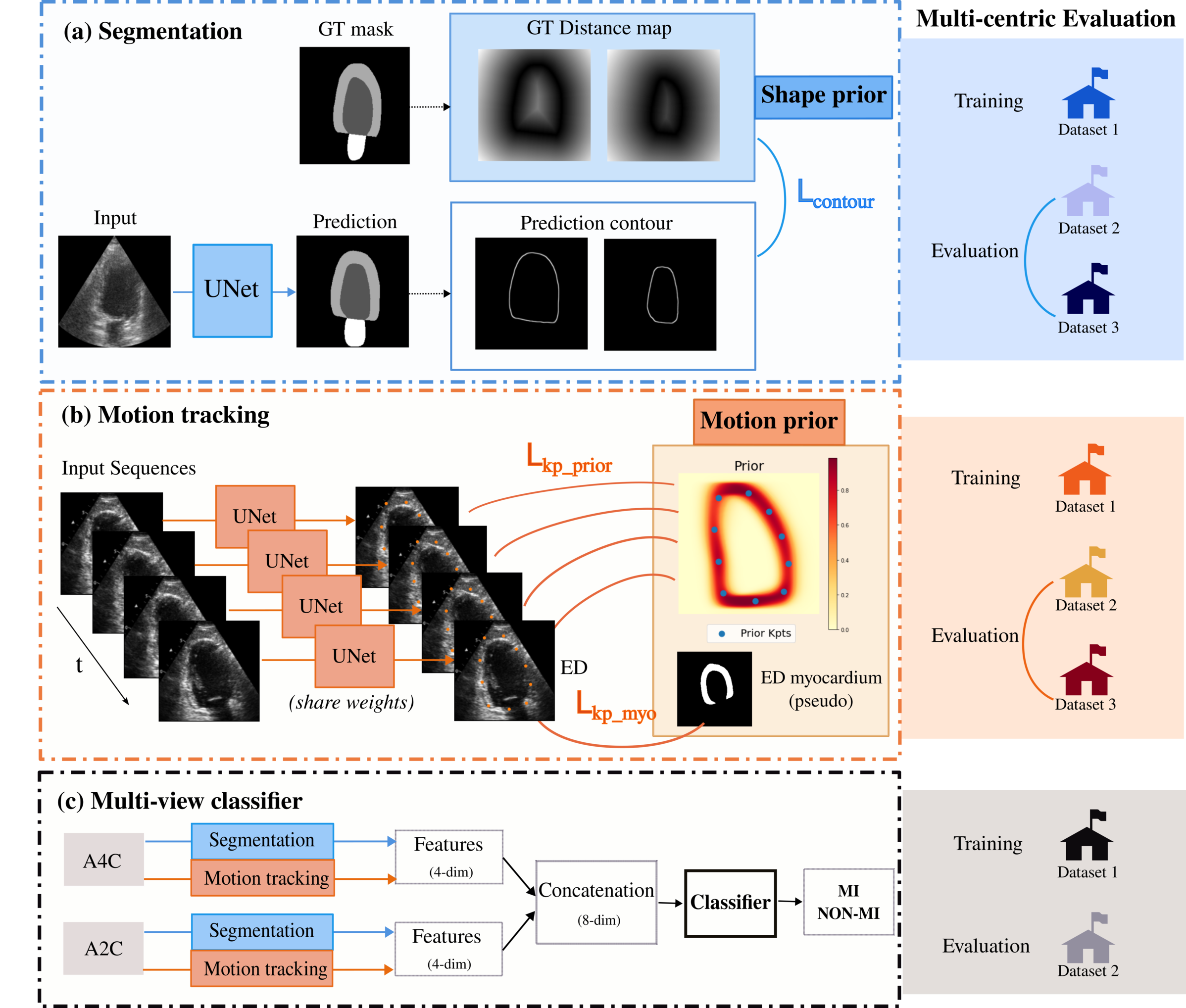
Echocardiography is a widely used medical modality to evaluate cardiac function. Automatic solutions for 2D echocardiography analysis could greatly influence cardiac disease diagnosis and prognosis. Within its diverse analysis tasks, segmentation and motion estimation are critical. Segmentation provides insights into cardiac structures, while motion estimation depicts myocardial deformation during cardiac cycles via image registration. These two tasks help extract useful clinical indexes, including ejection fraction and heart wall strain. Deep learning based methods that aim to tackle each of the above tasks have demonstrated efficient performance. However, the predominant focus on a single research dataset raises concerns about the generalisation of these approaches to other datasets and their applicability to downstream clinical tasks (diagnosis, prognosis...). In this paper, we propose a comprehensive echocardiography analysis pipeline, consisting of a segmentation module, a motion tracking module, and a classifier. The segmentation module incorporates a shape prior, leveraging a contour-aware loss function for the endocardium and epicardium. With appropriate data augmentation strategies, this module consistently demonstrates robust segmentation and ejection fraction estimation performance across diverse datasets from distinct medical centres. The motion tracking module integrates a motion prior for the myocardium through a differentiable poly-affine transformation, ensuring smooth and anatomy-aware deformation estimation. This module exhibits improved registration performance and maintains a more acceptable regularity in the deformation field across datasets. Finally, the classifier leverages features extracted from the preceding segmentation and motion tracking modules for downstream classification, providing interpretable inputs. The segmentation model and motion tracking model undergo individual training on two publicly available datasets. Subsequently, the classifier undergoes further training on a third pathology dataset using the estimated features from the previously trained models. To evaluate real-world performance, we directly apply the three trained models to a clinical dataset from a local hospital. Our successful application of these proposed methods in detecting patients with myocardial infarction (0.84 in F1 score and 0.77 in accuracy), a life-threatening cardiac disease, emphasises their potential for clinical utility. Our codes are available at https://github.com/xxx/xxx
Keywords
Image segmentation · Image registration · Generalisation of deep learning methods · priors · Echocardiography analysis
Bibtex
@article{melba:2024:031:yang,
title = "Echocardiography Analysis with Deep Learning using Priors: Multi-centric Evaluation of Generalisation",
author = "Yang, Yingyu and Rocher, Marie and Moceri, Pamela and Sermesant, Maxime",
journal = "Machine Learning for Biomedical Imaging",
volume = "2",
issue = "November 2024 issue",
year = "2024",
pages = "2293--2325",
issn = "2766-905X",
doi = "https://doi.org/10.59275/j.melba.2024-58fd",
url = "https://melba-journal.org/2024:031"
}
RIS
TY - JOUR
AU - Yang, Yingyu
AU - Rocher, Marie
AU - Moceri, Pamela
AU - Sermesant, Maxime
PY - 2024
TI - Echocardiography Analysis with Deep Learning using Priors: Multi-centric Evaluation of Generalisation
T2 - Machine Learning for Biomedical Imaging
VL - 2
IS - November 2024 issue
SP - 2293
EP - 2325
SN - 2766-905X
DO - https://doi.org/10.59275/j.melba.2024-58fd
UR - https://melba-journal.org/2024:031
ER -